
High intratumoral heterogeneity in clear cell renal cell carcinoma is associated with reduced immune response and survival
Highlight box
Key findings
• High-grade tumors had significantly higher mutant-allele tumor heterogeneity (MATH) values than low-grade tumors. Patients with lower MATH values had longer overall survival than those with higher MATH values, a result confirmed in a separate database of 106 clear cell renal cell carcinoma (ccRCC) patients.
• Lower MATH values correlated with increased infiltration of activated dendritic cells and reduced regulatory T cells in tumors compared to those with higher MATH values.
• MATH values hold potential as biomarkers for the stratification of ccRCC patients.
What is known and what is new?
• ccRCC is characterized by high intratumor heterogeneity (ITH), which is known to contribute to disease progression, metastasis, and resistance to various therapies.
• Our study aims to establish ITH as a clinically relevant biomarker for predicting outcomes and guiding treatment strategies in ccRCC, marking a novel approach in the field.
What is the implication, and what should change now?
• Our findings will most likely infer a useful biomarker for the prognostic evaluation and will facilitate the personalized management of this disease in patients.
Introduction
Renal cell carcinoma (RCC) is the seventh most frequently diagnosed type of malignant tumor (1). Clear cell renal cell carcinoma (ccRCC) is a most common histological subtype of RCC and it is associated with a broad range of clinical outcomes. Compared to other subtypes, ccRCC typically exhibits higher malignancy, a broader mutational landscape, elevated rates of metastasis and recurrence, and poorer clinical outcomes. Intratumor heterogeneity (ITH) serves as a critical factor in driving ccRCC progression and treatment resistance. ITH may be a limitation to developing precision medicine strategies based on gene mutation status and presents a significant challenge in the implementation of precision medicine (2).
Mutant-allele tumor heterogeneity (MATH) is a bioinformatics parameter providing a quantitative and measurable assessment of ITH (3,4). The prognostic influence of MATH has been studied in head and neck (3,4), breast (5), and colorectal cancer cohorts (6). Mroz et al. (3) observed a relationship between high MATH and worse overall survival in head and neck squamous cancers. McDonald et al. found that high MATH were associated with poor overall survival prognosis with estrogen receptor-positive and non-triple-negative breast cancers (5). Rajput et al. (6) found that higher MATH correlated strongly with the risk of colon cancer metastases. Together, the above suggest that ITH is associated with poor prognosis and has prognostic relevance in some tumors.
Despite recent advancements in systemic therapies for advanced ccRCC, including tyrosine kinase inhibitors, mTOR inhibitors, and immune checkpoint inhibitors targeting PD-1/cytotoxic T-lymphocyte-associated protein 4 (CTLA-4) (7-9), resistance mechanisms and immune evasion remain critical challenges. Emerging evidence indicates that the interplay between regulatory T cells (Tregs) and TGF-β signaling within the tumor microenvironment may contribute to treatment resistance, highlighting the need to better understand the underlying molecular and immunological mechanisms (10,11). The extent of heterogeneity in ccRCC remains unknown, and currently, there is a lack of clinically established approaches to leverage ITH for prognostic prediction in patients. Therefore, the objective of this study was to evaluate ITH in ccRCC as a potential predictor of clinical outcomes. The Bioconductor R package Maftools was utilized to conduct a thorough and efficient analysis of somatic variants in ccRCC in The Cancer Genome Atlas (TCGA). Then MATH algorithm was used to quantify ITH in ccRCC and explore its relationship with clinical characteristics. We demonstrated MATH value as an independent prognostic marker of ccRCC patients, a finding that was verified in another independent ccRCC database. However, although genomic data reveal the extent of genetic heterogeneity within ccRCC, the possible factors associated with tumor heterogeneity and patient outcome are unclear. As tumors grow, some mutations produce neoantigens that affect the patient’s response to immune checkpoint inhibitors. It has been found that tumors with high heterogeneity may produce neoantigens that recruit immune cells, which may exert selective pressure to form tumor heterogeneity (12,13).
The prognostic significance of tumor-infiltrating lymphocytes (TILs) in multiple human tumors has revealed a significant association with patient survival (14-16). However, specific subsets of T lymphocytes are critically involved in shaping the immunological landscape of the tumor microenvironment. Cytotoxic (CD8+) T cells are key effectors in antitumor immunity, and elevated levels of CD8+ T cell infiltration have been demonstrated to correlate with favorable prognostic outcomes (17). Some studies suggest that Tregs are capable of suppressing antitumor immune responses in the tumor microenvironment through the inhibition of cytotoxic T cell activity (18,19). The antitumor activity of TILs can be inhibited by Tregs expressing forkhead box P3 (FoxP3), and the composition and proportion of TILs have stronger prognostic implications in tumor microenvironment (20,21). FoxP3 is an important transcription factor for the function and development of Tregs (22,23), and it is considered to be a specific Tregs marker. FoxP3+ Tregs are essential suppressors that function in maintaining immunological tolerance to host-derived tissues (24). Increased infiltration of FoxP3+ Tregs in the tumor microenvironment is linked to immunosuppression and poor prognosis (25). Thus, FoxP3+ Tregs are emerging as a promising prognostic factor and therapeutic target in the context of ccRCC. TGF-β is a central mediator in the regulation of T cell-driven immune responses and the establishment of immune tolerance (26,27). Research has shown that TGF-β induces the conversion of naive CD4+CD25− T cells into CD4+CD25+ Tregs by upregulating FoxP3 expression (28). In this ccRCC cohort, the relationship between ITH and immunogenicity landscape has not been fully explored. Therefore, a MATH algorithm was used to measure ITH and a systematic analysis of ccRCC patient survival outcomes based on MATH value was conducted. Using TCGA, we explored the correlation of MATH value with clinical parameters, infiltrating immune cell compositions, and immune response genes. We present this article in accordance with the REMARK reporting checklist (available at https://tau.amegroups.com/article/view/10.21037/tau-2024-741/rc).
Methods
Patients and clinical parameters
The clinical data analyzed in this study were publicly accessible through the TCGA Data Portal (https://portal.gdc.cancer.gov) and were downloaded from cBioPortal for further analysis. The study was conducted in accordance with the Declaration of Helsinki and its subsequent amendments. Independent ccRCC cohorts for validating were selected based on the following criteria: (I) high-quality RNA sequencing data; (II) comprehensive clinical annotations; (III) sufficient sample size for statistical analysis; (IV) consistency in analytical pipelines to minimize batch effects; and (V) relevance to study objectives. Finally, an independent cohort of mutation and clinical data was retrieved from cBioPortal to validate the results (29,30).
Analysis of genomic variants and MATH score computation
ccRCC somatic variants were acquired in mutation annotation format (MAF) from TCGA. We used Maftools, an R Bioconductor package (31), to assess somatic variants in ccRCC.
For each ccRCC sample, the MATH value was calculated based on the protocol established in earlier research (3,4). The process of calculating the MATH value consists of the following steps: (I) determining the mutated allele fraction (MAF) for each locus, defined as the ratio of mutated reads to the total number of reads; (II) compute the absolute deviation of each MAF relative to the median MAF value, followed by multiplying the median of these deviations by a scaling constant (1.4826) to yield the median absolute deviation (MAD); (III) calculate the ratio of the MAD to the median MAF of the tumor’s mutated genomic loci, and express this value as a percentage using the formula: MATH = 100 * MAD/median.
Immune cell fraction analysis
Immune cell compositions were determined using cell-type identification by estimating relative subsets of RNA transcripts (CIBERSORT), a bioinformatic algorithm that calculates immune cell compositions from tumor gene expression profiles (32). The immune cell fraction data was downloaded through The Cancer Immunome Atlas (TCIA) (https://tcia.at/home) (33). Each immune cell fraction was compared between low-MATH and high-MATH ccRCC samples.
Statistical analysis
The Kolmogorov-Smirnov test was used to analyze the distribution characteristics of the data. The correlations between MATH values and clinical characteristics were evaluated using the Kruskal-Wallis test. Prognostic assessments were carried out using Kaplan-Meier survival analyses, with statistical significance assessed by log-rank tests. All statistical procedures were performed using the R packages “survminer” and “survival” (R version 4.3.0), and a P value <0.05 was deemed statistically significant.
Results
Clinical characteristics of patients with ccRCC
We used Maftools to analyze genome sequencing data obtained from Data Portal of TCGA ccRCC. The demographic, clinical, and pathological characteristics of all patients included in the present analysis are outlined in Table 1. The MATH algorithm was applied to evaluate ITH, and its relationship with clinical parameters was systematically analyzed. We found that MATH was significantly correlated with the grade of ccRCC (P<0.05, Table 1).
Table 1
| Characteristics | Number of patients | Percent (%) | MATH ± SD | P value |
|---|---|---|---|---|
| Age (years) | 0.53 | |||
| 26–50 | 72 | 21.4 | 27.78±14 | |
| 51–70 | 189 | 56.3 | 30.3±15 | |
| 71–90 | 75 | 22.3 | 31.0±17 | |
| Gender | 0.88 | |||
| Male | 216 | 64.3 | 31.1±15 | |
| Female | 120 | 35.7 | 28.5±16 | |
| Race | 0.65 | |||
| Asian | 6 | 1.8 | 28.9±18 | |
| Caucasian | 272 | 82.4 | 29.8±15 | |
| African | 52 | 15.8 | 31.1±15 | |
| Unknown | 6 | – | – | |
| Stage | 0.050 | |||
| Stage-I | 192 | 57.5 | 28.2±15 | |
| Stage-II | 33 | 9.9 | 30.2±18 | |
| Stage-III | 68 | 20.4 | 30.8±14 | |
| Stage-IV | 41 | 12.3 | 34.4±17 | |
| Unknown | 2 | – | – | |
| Lymph node stage | 0.49 | |||
| N0 | 139 | 93.9 | 29.1±17 | |
| N1 | 9 | 6.1 | 31.3±8 | |
| Unknown | 188 | – | – | |
| Metastasis stage | 0.08 | |||
| M0 | 267 | 87.8 | 29.3±16 | |
| M1 | 37 | 12.2 | 34.4±12 | |
| Unknown | 32 | – | – | |
| Histologic grade | **0.001 | |||
| Grade 1 | 13 | 4.0 | 27.4±10 | |
| Grade 2 | 150 | 45.6 | 27.8±15 | |
| Grade 3 | 123 | 37.4 | 30.3±13 | |
| Grade 4 | 43 | 13.1 | 37.4±17 | |
| Unknown | 7 | – | – | |
| Recurrence | 0.16 | |||
| Recurrence | 70 | 24.0 | 32.1±17 | |
| Free | 222 | 76.0 | 29.7±15 | |
| Unknown | 44 | – | – | |
**. ccRCC, clear cell renal cell carcinoma; MATH, mutant-allele tumor heterogeneity; SD, standard deviation; TCGA, The Cancer Genome Atlas.
Comprehensive analysis of somatic variants in ccRCC
Somatic variants in ccRCC were analyzed using the Bioconductor R package Maftools, enabling efficient and comprehensive processing of TCGA-derived data. We used the Maftools function oncodrive, which identifies cancer genes (driver) from a given MAF and is based on the algorithm oncodriveCLUST (34).
Maftools analysis revealed 18,310 somatic mutations, consisting of 12,997 missense mutations, 1,259 nonsense mutations, 1,551 insertions, 1,970 deletions, 25 translation start site variants, 490 splice site variants, and 18 nonstop mutations (Figure 1A).
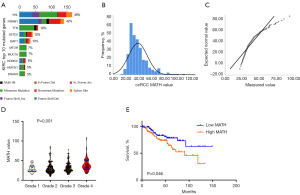
MATH value characteristics and their relationship with clinical factors
ccRCC displays high ITH during its progression. ITH can be assessed using a MATH value (4), which reflects an aspect of tumor biology distinct from mutation rate and has been shown to be a simple, quantitative, and broadly applicable strategy for assessing ITH. In this analysis, we quantified MATH values in the TCGA ccRCC cohort to evaluate the clinical impact of ITH in ccRCC. Figure 1B displays the distribution of MATH values. The quantile-quantile (Q-Q) plot (Figure 1C) and Kolmogorov-Smirnov test (P<0.001) provide evidence that MATH values in this cohort significantly deviate from a normal distribution. By applying a rank-sum test of variance, we observed significant relationships between MATH values and tumor grade (Figure 1D, P=0.001) as well as tumor stage (Table 1, P=0.050). A significant increase in MATH values was observed in the high-grade group compared to the low-grade group (P<0.005), suggesting that MATH values could be a valuable tool for distinguishing between these patient cohorts. The interquartile range of MATH values spanned from 23.6 (lower quartile) to 39.4 (upper quartile). Cases with MATH values <23.6 were classified into the “low-MATH” group, and cases with a MATH value >39.4 were classified into the “high-MATH” group. We compared the proportion of Grade 4 cases in the high-MATH versus the low-MATH. Although the proportion of Grade 4 cases was higher in the higher high-MATH in the high-MATH (17.86% vs. 6%), this difference did not reach statistical significance (P=0.12). To investigate the prognostic implications of MATH values, we analyzed their significance in the low- and high-MATH groups. MATH value was independently associated with overall survival in the ccRCC cohort (P=0.046, Figure 1E). A high MATH value was associated with worse survival in ccRCC. Taken together, we found that a higher MATH value is not only related to high tumor grade but is also an independent risk factor for reduced overall survival in ccRCC patients.
Validation of MATH value characteristics and their relationship with clinical grade and prognosis in another independent ccRCC cohort
To further investigate whether MATH value can be widely applied to predict the outcome of ccRCC patients, we investigated another ccRCC cohort, which was composed of 106 ccRCC patients and was published in 2013 (29). We acquired comprehensive mutation and clinical data from cBioPortal and conducted an analysis to determine their prognostic utility.
First, we calculated MATH values in this independent ccRCC cohort. Figure 2A illustrates the distribution of MATH values. The Q-Q plot (Figure 2B) and Kolmogorov-Smirnov test (P<0.001) revealed that the distribution of MATH values from this database was consistent with that of the TCGA ccRCC cohort. Through a rank-sum test of variance, we analyzed the overall trend of median MATH values and found that the high-grade group exhibited higher values than the low-grade group. However, this difference was not statistically significant (Figure 2C, P=0.11), possibly due to the limited sample size in this cohort. Next, we investigated the prognostic significance of MATH value in this independent ccRCC cohort. The MATH values exhibited a lower quartile of 22.4 and an upper quartile of 42.5. Thus, samples with MATH values <22.4 were classified into the “low-MATH” group, and cases with a MATH value >42.5 were classified into the “high-MATH” group. We identified a trend toward improved survival time in the low-MATH group compared to the high-MATH group (Figure 2D, P=0.08). However, this difference was not statistically significant, possibly due to the small cohort size in these groups. Taken together, our data show that low MATH values demonstrated favorable prognostic significance in a second, independent ccRCC cohort.

ccRCC with high MATH value are associated with reduced immune cell infiltration and increased immunosuppressive Tregs
The relationship between immune cell infiltration and prognosis and survival has been reported in a wide range of human tumors. However, whether the prognosis of patients with low- and high-MATH ccRCC is related to immune cell infiltration is currently unknown. Therefore, we evaluated the composition of immune cells in the ccRCC cohort and investigated its relationship with ITH. Immune cell fractions in the high- and low-MATH ccRCC groups were compared using CIBERSORT. Our statistical analyses reveal no significant differences between low- and high-MATH ccRCC in their fraction of activated memory CD4 T cells (Figure 3A, P=0.91) and CD8+ T cells (Figure 3B, P=0.39), but high-MATH ccRCC were associated with reduced proportions of activated dendritic cells (DCs) (Figure 3C, P=0.048), which play antitumor roles in ccRCC. Interestingly, compared with low-MATH tumors, high-MATH tumors were also associated with significantly higher fractions of immunosuppressive Tregs (Figure 3D, P=0.02). We found that tumors with high MATH displayed reduced antitumor immune cells and increased immunosuppressive Tregs, which is consistent with the view that ITH is a multiple contributing factor and immune cell infiltration is a vital factor. This finding further supports the notion that reduced immune cell infiltrations allow the tumor to clonally evolve and thus develop high ITH with high MATH.
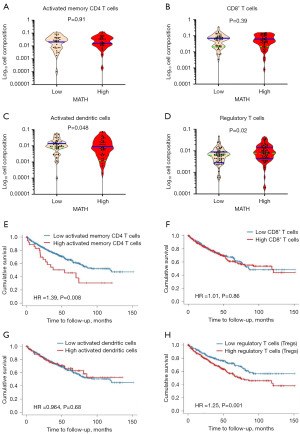
Overall survival of ccRCC decreased with increased immunosuppressive Tregs infiltration
To further elucidate the immune cell infiltration effect on the survival and prognosis of ccRCC patients, we analyzed the survival and prognosis of 533 ccRCC patients in the TCGA database by Tumor Immune Estimation Resource (TIMER) 2.0 (35-37). Patients were divided into two groups according to the composition of immune cell infiltration, and the results showed that CD8+ T cells and activated DCs had no significant effect on the survival prognosis of patients (Figure 3E, P=0.86; Figure 3F, P=0.68), but high activated memory CD4 T cells and increased immunosuppressive Tregs significantly reduced the overall survival time of patients (Figure 3G, P=0.008; Figure 3H, P=0.001). Tregs are a component of the immune system that can suppress immune responses by affecting the activation of other cell types. Since Tregs are involved in cancer immune evasion and escape and therefore contribute to tumor development and progression, which suggested it was a significant risk factor for reducing the overall survival of ccRCC patients.
ccRCC with high MATH are associated with increased FoxP3 and TGF-β expression
Immune checkpoint inhibitors provide a model for cancer treatment and most of the immune checkpoint molecules are expressed in Tregs. Immune checkpoint molecules such as CTLA4, PD-L1, and PD-1, also known as T cell exhaustion markers, often reflect the immunogenicity of tumors. The expression of these checkpoint molecules normally inhibits T cell function and may be exploited during immune escape in tumor tissue. Although no significant difference was found in the composition of activated memory CD4+ and CD8+ T cells between low- and high-MATH ccRCC (Figure 3A,3E), whether tumor heterogeneity affects the expression of T cell exhaustion markers is unclear. Therefore, we compared the expressions of PDCD1, CD274, and CTLA4 in low- and high-MATH ccRCC. We found that low- and high-MATH ccRCC were not associated with expression of T-cell exhaustion markers PDCD1 (Figure 4A, P=0.23), CTLA4 (Figure 4B, P=0.35), and CD274 (Figure 4C, P=0.84). This result is consistent with the above finding that ccRCC with low and high MATH are not associated with active T cell infiltration.
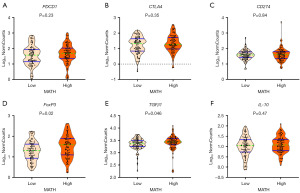
FoxP3 is an important transcription factor for the function and development of Tregs and is considered the most specific Treg marker. Interleukin-10 (IL-10) and TGF-β are critical factors in regulating T cell-mediated immune responses. Thus, we compared the expressions of FoxP3, TGF-β1 and IL-10 in low- and high-MATH ccRCC. Interestingly, compared with low-MATH ccRCC, high-MATH group were significantly correlated with higher expression of FoxP3 (Figure 4D, P=0.02) and TGF-β1 (Figure 4E, P=0.046) but not with IL-10 expression (Figure 4F, P=0.47).
A total of 533 ccRCC patients were found in TCGA database. Wilcoxon test analysis by TMIER2.0 showed that FoxP3 and TGF-β1 expression levels in tumor tissues were significantly higher than normal tissues (Figure 5A, P<0.001; Figure 5B, P<0.001). Overall survival analysis showed that patients with high FoxP3 or TGF-β1 expression had significantly lower overall survival than those with low FoxP3 or TGF-β1 expression (Figure 5C, P<0.05; Figure 5D, P<0.001). Further Spearman’s correlation analysis showed that FoxP3 and TGF-β1 expression levels were significantly correlated with Tregs infiltration degree (Figure 5E, P<0.001; Figure 5F, P<0.001), as well as FoxP3 and TGF-β1 expression levels were also significantly positively correlated (Figure 5G, P<0.001). These results suggested that FoxP3+ Tregs realized their immune tolerance in high-MATH ccRCC through TGF-β1 regulation of FoxP3 expression. FoxP3+ Tregs are a potential prognostic factor and therapeutic target.

Discussion
ccRCC is a most common histological subtype of RCC which is characterized by a more diverse mutation spectrum, higher overall mutation rates, and worse overall prognosis. Similar to most solid tumors, ccRCC is spatially heterogeneous. ITH is crucial in promoting the progression of ccRCC and its resistance to various treatments. The significant genetic diversity within tumors is believed to increase the risk of poor prognosis and reduced survival rates for patients suffering from ccRCC.
Calculation of MATH value is a novel method to quantify ITH. Several previous studies reported use of MATH value as a prognostic biomarker for patients with squamous cell carcinoma of head and neck (3,4), breast cancer (5), and colorectal cancer (6). However, the use of MATH values to evaluate ITH of ccRCC has not been fully explored at the genomic level. Novel biomarkers could provide more information on patients’ prognosis with ccRCC (38,39). Our study represents the first comprehensive investigation into the prognostic significance of MATH in ccRCC. ITH accounts for the differences in tumor behavior among different patients, and in our study, we found that high MATH value was not only related to high tumor grade but was also an independent risk factor for overall survival.
Our findings demonstrate a significant stepwise increase in MATH values with ascending histologic grade, highlighting the critical role of ITH in ccRCC progression. This trend aligns with the well-established association between high-grade tumors and genomic instability, which drives clonal diversification and aggressive behavior (40). The increase in MATH with both stage (P=0.050) and grade (P<0.001) highlight a unified axis of tumor progression. Advanced-stage and high-grade tumors likely share common drivers of heterogeneity, such as chromosomal instability and subclonal evolution, which facilitate immune evasion and metastasis (40-42). Despite the borderline P value (potentially limited by the smaller M1 subgroup, n=37), the elevated MATH in M1 tumors suggests that extreme heterogeneity may drive metastasis by selecting for aggressive clones resistant to immune surveillance.
The relationship between ITH and tumor immunity is a controversial issue. Some researchers propose that tumor heterogeneity generates neoantigens, which can attract immune cells and potentially enhance anti-tumor immune responses. In contrast, others argue that the presence and activity of immune cells exert selective pressure on tumors, thereby shaping and controlling tumor heterogeneity (13,43). We aimed to explore the association between immune cell infiltration and ITH. However, different types of immune cells may play different roles in the tumor microenvironment. Some studies have shown that Tregs can inhibit tumor immunity in the tumor microenvironment (44). An increased proportion of FoxP3+ Tregs is associated with poor prognosis for glioblastoma, breast cancer (45), colorectal cancer (46), gastric cancer (47), and cervical cancer (48). TGF-β may contribute to increases in FoxP3+ cells; thus, FoxP3+ Tregs represent a potential prognostic factor and therapeutic target (46). Interestingly, our study shown that high-MATH tumors were correlated with significantly higher proportions of immunosuppressive Tregs compared with low-MATH tumors (Figure 3D, P=0.02). TGF-β and IL-10 are both critical factors in regulation of T cell-mediated immune responses (49). Compared with low-MATH tumors, we found a significant correlation between higher expression of FoxP3 (Figure 4D, P=0.02) and TGF-β (Figure 4E, P=0.046) with high MATH and but not with IL-10 expression (Figure 4F, P=0.47). These results suggested that FoxP3+ Tregs realize their immune tolerance in high-MATH ccRCC through TGF-β regulation of Foxp3 expression. FoxP3+ Tregs should be investigated as a potential prognostic factor and therapeutic target in ccRCC patients.
DCs are the most effective antigen presenting cells and are essential for initiating the host’s defensive immune responses (50,51). DCs are important components of the tumor microenvironment and may play crucial roles in tumorigenesis, tumor progression, and metastasis. As one of the most potent immunosuppressive cytokines, TGF-β is important for malignant tumor progression (52,53). TGF-β, a tumor-derived factor, is recognized for its ability to impede DC maturation by downregulating the expression of CD1d, a surface marker critical for antigen presentation. Beyond suppressing DC maturation, tumor-derived TGF-β also promotes DC apoptosis, further compromising the immune system’s ability to mount an effective anti-tumor response (52,54). Our data show that high-MATH tumors were associated with significantly lower fractions of activated DCs (Figure 3B, P=0.048) and higher TGF-β1 expression (Figure 4E, P=0.046). These results suggested that the correlation of high MATH with worse survival may be due to TGF-β inhibition of DC maturation.
Our findings highlight the FoxP3/TGF-β axis as a key regulator of immune suppression in ccRCC, offering potential for novel therapeutic strategies, such as combining TGF-β inhibitors with immune checkpoint blockade to enhance anti-tumor immunity. However, significant knowledge gaps remain, particularly regarding the mechanism by which ITH influences immune infiltration, which is not yet fully understood. Due to statistical power constraints and dataset limitations, subgroup analyses were not explored. Future research should leverage multi-omics approaches and single-cell technologies to unravel these complexities, while clinical trials will be essential to validate therapeutic targets. Over the next 5 years, we anticipate advancements in personalized immunotherapies and predictive biomarkers, ultimately improving outcomes for ccRCC patients.
MATH value renders a quantifiable estimate for ITH and is less constrained by somatic mutation number and sequencing depth in the ccRCC cohorts. The use of MATH value to quantify ITH is a valuable method, however, the standards and algorithm are different for each somatic caller in different databases, so the results vary between databases. Therefore, utilizing of MATH value to quantify heterogeneity of tumor requires adoption of universal standards for calling somatic mutations. However, ITH as represented by MATH may be confused by the number of somatic copy number alterations, which should not be ignored.
There are several potential limitations in this study, due to the nature of available ccRCC cohorts. First, information regarding pre- and postoperative treatment is not available for the majority of the patients with ccRCC, and thus, how these treatments impacted overall survival could not be fully considered in our analyses. Moreover, the majority of ccRCC patients in our study were Caucasian, which limits the generalizability of our findings to other populations. Furthermore, despite adequate sample size, the non-uniformity of patient follow-up data constrained our clinical correlation analysis. Therefore, future studies should incorporate samples from diverse racial groups. There are several potential limitations in this study, to validate our findings. Additionally, further validation using a variety of algorithms is also necessary.
Conclusions
In our research, we conducted a systematic analysis of ccRCC patients’ survival outcomes based on ITH and found that MATH is a useful biomarker for identifying patients with ccRCC. High MATH correlates with decreased immune response and worse survival in ccRCC patients.
Acknowledgments
We would like to thank “TCGA Research Network” for generating, curating and providing ccRCC clinical data, the results of this study are based on data generated by TCGA.
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at https://tau.amegroups.com/article/view/10.21037/tau-2024-741/rc
Peer Review File: Available at https://tau.amegroups.com/article/view/10.21037/tau-2024-741/prf
Funding: This work was supported by
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tau.amegroups.com/article/view/10.21037/tau-2024-741/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki and its subsequent amendments.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Znaor A, Lortet-Tieulent J, Laversanne M, et al. International variations and trends in renal cell carcinoma incidence and mortality. Eur Urol 2015;67:519-30. [Crossref] [PubMed]
- Thole TM, Toedling J, Sprüssel A, et al. Reflection of neuroblastoma intratumor heterogeneity in the new OHC-NB1 disease model. Int J Cancer 2020;146:1031-41. [Crossref] [PubMed]
- Mroz EA, Tward AD, Hammon RJ, et al. Intra-tumor genetic heterogeneity and mortality in head and neck cancer: analysis of data from the Cancer Genome Atlas. PLoS Med 2015;12:e1001786. [Crossref] [PubMed]
- Mroz EA, Rocco JW. MATH, a novel measure of intratumor genetic heterogeneity, is high in poor-outcome classes of head and neck squamous cell carcinoma. Oral Oncol 2013;49:211-5. [Crossref] [PubMed]
- McDonald KA, Kawaguchi T, Qi Q, et al. Tumor Heterogeneity Correlates with Less Immune Response and Worse Survival in Breast Cancer Patients. Ann Surg Oncol 2019;26:2191-9. [Crossref] [PubMed]
- Rajput A, Bocklage T, Greenbaum A, et al. Mutant-Allele Tumor Heterogeneity Scores Correlate With Risk of Metastases in Colon Cancer. Clin Colorectal Cancer 2017;16:e165-70. [Crossref] [PubMed]
- Rizzo A, Mollica V, Marchetti A, et al. Adjuvant PD-1 and PD-L1 Inhibitors and Relapse-Free Survival in Cancer Patients: The MOUSEION-04 Study. Cancers (Basel) 2022;14:4142. [Crossref] [PubMed]
- Santoni M, Buti S, Myint ZW, et al. Real-world Outcome of Patients with Advanced Renal Cell Carcinoma and Intermediate- or Poor-risk International Metastatic Renal Cell Carcinoma Database Consortium Criteria Treated by Immune-oncology Combinations: Differential Effectiveness by Risk Group? Eur Urol Oncol 2024;7:102-11. [Crossref] [PubMed]
- Santoni M, Mollica V, Rizzo A, et al. Dynamics of resistance to immunotherapy and TKI in patients with advanced renal cell carcinoma. Cancer Treat Rev 2025;133:102881. [Crossref] [PubMed]
- Marchetti A, Rosellini M, Mollica V, et al. The Molecular Characteristics of Non-Clear Cell Renal Cell Carcinoma: What's the Story Morning Glory? Int J Mol Sci 2021;22:6237. [Crossref] [PubMed]
- Rosellini M, Marchetti A, Mollica V, et al. Prognostic and predictive biomarkers for immunotherapy in advanced renal cell carcinoma. Nat Rev Urol 2023;20:133-57. [Crossref] [PubMed]
- Black JRM, McGranahan N. Genetic and non-genetic clonal diversity in cancer evolution. Nat Rev Cancer 2021;21:379-92. [Crossref] [PubMed]
- McGranahan N, Furness AJ, Rosenthal R, et al. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science 2016;351:1463-9. [Crossref] [PubMed]
- Ferkel SAM, Holman EA, Sojwal RS, et al. Tumor-Infiltrating Immune Cells in Colorectal Cancer. Neoplasia 2025;59:101091. [Crossref] [PubMed]
- Brummel K, Eerkens AL, de Bruyn M, et al. Tumour-infiltrating lymphocytes: from prognosis to treatment selection. Br J Cancer 2023;128:451-8. [Crossref] [PubMed]
- Bai YG, Gao GX, Zhang H, et al. Prognostic value of tumor-infiltrating lymphocyte subtypes in residual tumors of patients with triple-negative breast cancer after neoadjuvant chemotherapy. Chin Med J (Engl) 2020;133:552-60. [Crossref] [PubMed]
- Raskov H, Orhan A, Christensen JP, et al. Cytotoxic CD8(+) T cells in cancer and cancer immunotherapy. Br J Cancer 2021;124:359-67. [Crossref] [PubMed]
- Zhang A, Fan T, Liu Y, et al. Regulatory T cells in immune checkpoint blockade antitumor therapy. Mol Cancer 2024;23:251. [Crossref] [PubMed]
- Liang Y, Qiao L, Qian Q, et al. Integrated single-cell and spatial transcriptomic profiling reveals that CD177(+) Tregs enhance immunosuppression through apoptosis and resistance to immunotherapy in hepatocellular carcinoma. Oncogene 2025;44:1578-91. [Crossref] [PubMed]
- Whiteside TL. FOXP3+ Treg as a therapeutic target for promoting anti-tumor immunity. Expert Opin Ther Targets 2018;22:353-63. [Crossref] [PubMed]
- Khattri R, Cox T, Yasayko SA, et al. An essential role for Scurfin in CD4+CD25+ T regulatory cells. Nat Immunol 2003;4:337-42. [Crossref] [PubMed]
- Deng G, Song X, Greene MI. FoxP3 in T(reg) cell biology: a molecular and structural perspective. Clin Exp Immunol 2020;199:255-62. [Crossref] [PubMed]
- Mortezaee K. FOXP3 (in)stability and cancer immunotherapy. Cytokine 2024;178:156589. [Crossref] [PubMed]
- Sakaguchi S, Yamaguchi T, Nomura T, et al. Regulatory T cells and immune tolerance. Cell 2008;133:775-87. [Crossref] [PubMed]
- Li Y, Zhang C, Jiang A, et al. Potential anti-tumor effects of regulatory T cells in the tumor microenvironment: a review. J Transl Med 2024;22:293. [Crossref] [PubMed]
- Chen W. TGF-β Regulation of T Cells. Annu Rev Immunol 2023;41:483-512. [Crossref] [PubMed]
- Koh J, Hur JY, Lee KY, et al. Regulatory (FoxP3(+)) T cells and TGF-β predict the response to anti-PD-1 immunotherapy in patients with non-small cell lung cancer. Sci Rep 2020;10:18994. [Crossref] [PubMed]
- Chen W, Jin W, Hardegen N, et al. Conversion of peripheral CD4+CD25- naive T cells to CD4+CD25+ regulatory T cells by TGF-beta induction of transcription factor Foxp3. J Exp Med 2003;198:1875-86. [Crossref] [PubMed]
- Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:pl1. [Crossref] [PubMed]
- Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012;2:401-4. [Crossref] [PubMed]
- Mayakonda A, Lin DC, Assenov Y, et al. Maftools: efficient and comprehensive analysis of somatic variants in cancer. Genome Res 2018;28:1747-56. [Crossref] [PubMed]
- Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods 2015;12:453-7. [Crossref] [PubMed]
- Charoentong P, Finotello F, Angelova M, et al. Pan-cancer Immunogenomic Analyses Reveal Genotype-Immunophenotype Relationships and Predictors of Response to Checkpoint Blockade. Cell Rep 2017;18:248-62. [Crossref] [PubMed]
- Tamborero D, Gonzalez-Perez A, Lopez-Bigas N. OncodriveCLUST: exploiting the positional clustering of somatic mutations to identify cancer genes. Bioinformatics 2013;29:2238-44. [Crossref] [PubMed]
- Li T, Fu J, Zeng Z, et al. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res 2020;48:W509-14. [Crossref] [PubMed]
- Li T, Fan J, Wang B, et al. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res 2017;77:e108-10. [Crossref] [PubMed]
- Arora M, Kumari S, Singh J, et al. Expression pattern, regulation, and clinical significance of TOX in breast cancer. Cancer Immunol Immunother 2021;70:349-63. [Crossref] [PubMed]
- Chen L, Luo Y, Wang G, et al. Prognostic value of a gene signature in clear cell renal cell carcinoma. J Cell Physiol 2019;234:10324-35. [Crossref] [PubMed]
- Zeng MH, Qiu JG, Xu Y, et al. IDUA, NDST1, SAP30L, CRYBA4, and SI as novel prognostic signatures clear cell renal cell carcinoma. J Cell Physiol 2019;234:16320-7. [Crossref] [PubMed]
- Li Y, Lih TM, Dhanasekaran SM, et al. Histopathologic and proteogenomic heterogeneity reveals features of clear cell renal cell carcinoma aggressiveness. Cancer Cell 2023;41:139-163.e17. [Crossref] [PubMed]
- Ran X, Xiao J, Zhang Y, et al. Low intratumor heterogeneity correlates with increased response to PD-1 blockade in renal cell carcinoma. Ther Adv Med Oncol 2020;12:1758835920977117. [Crossref] [PubMed]
- Golkaram M, Kuo F, Gupta S, et al. Spatiotemporal evolution of the clear cell renal cell carcinoma microenvironment links intra-tumoral heterogeneity to immune escape. Genome Med 2022;14:143. [Crossref] [PubMed]
- McGranahan N, Swanton C. Clonal Heterogeneity and Tumor Evolution: Past, Present, and the Future. Cell 2017;168:613-28. [Crossref] [PubMed]
- deLeeuw RJ, Kost SE, Kakal JA, et al. The prognostic value of FoxP3+ tumor-infiltrating lymphocytes in cancer: a critical review of the literature. Clin Cancer Res 2012;18:3022-9. [Crossref] [PubMed]
- Zhou Y, Shao N, Aierken N, et al. Prognostic value of tumor-infiltrating Foxp3+ regulatory T cells in patients with breast cancer: a meta-analysis. J Cancer 2017;8:4098-105. [Crossref] [PubMed]
- Suzuki H, Chikazawa N, Tasaka T, et al. Intratumoral CD8(+) T/FOXP3 (+) cell ratio is a predictive marker for survival in patients with colorectal cancer. Cancer Immunol Immunother 2010;59:653-61. [Crossref] [PubMed]
- Nagase H, Takeoka T, Urakawa S, et al. ICOS(+) Foxp3(+) TILs in gastric cancer are prognostic markers and effector regulatory T cells associated with Helicobacter pylori. Int J Cancer 2017;140:686-95. [Crossref] [PubMed]
- Shah W, Yan X, Jing L, et al. A reversed CD4/CD8 ratio of tumor-infiltrating lymphocytes and a high percentage of CD4(+)FOXP3(+) regulatory T cells are significantly associated with clinical outcome in squamous cell carcinoma of the cervix. Cell Mol Immunol 2011;8:59-66. [Crossref] [PubMed]
- Rouas R, Merimi M, Najar M, et al. Human CD8(+) CD25 (+) CD127 (low) regulatory T cells: microRNA signature and impact on TGF-β and IL-10 expression. J Cell Physiol 2019;234:17459-72. [Crossref] [PubMed]
- Wallet MA, Sen P, Tisch R. Immunoregulation of dendritic cells. Clin Med Res 2005;3:166-75. [Crossref] [PubMed]
- Zanna MY, Yasmin AR, Omar AR, et al. Review of Dendritic Cells, Their Role in Clinical Immunology, and Distribution in Various Animal Species. Int J Mol Sci 2021;22:8044. [Crossref] [PubMed]
- Liu CC, Wang YS, Lin CY, et al. Transient downregulation of monocyte-derived dendritic-cell differentiation, function, and survival during tumoral progression and regression in an in vivo canine model of transmissible venereal tumor. Cancer Immunol Immunother 2008;57:479-91. [Crossref] [PubMed]
- Fogel-Petrovic M, Long JA, Misso NL, et al. Physiological concentrations of transforming growth factor beta1 selectively inhibit human dendritic cell function. Int Immunopharmacol 2007;7:1924-33. [Crossref] [PubMed]
- Ito M, Minamiya Y, Kawai H, et al. Tumor-derived TGFbeta-1 induces dendritic cell apoptosis in the sentinel lymph node. J Immunol 2006;176:5637-43. [Crossref] [PubMed]


