
Clinical significance of urine-based automated detection of GSTP1 methylation for the diagnosis of suspected prostate cancer patients
Highlight box
Key findings
• Urinary glutathione S-transferase Pi-1 (GSTP1) methylation demonstrated higher specificity (83.6%) and accuracy (82.5%) compared to prostate-specific antigen (PSA; 54.1% and 60.8%, respectively).
• The combined method of urinary GSTP1 methylation and PSA achieved an area under the curve of 0.89, significantly enhancing diagnostic efficacy.
• In the PSA gray zone (4.0–10.0 ng/mL), GSTP1 methylation showed sensitivity and specificity of 87.5% and 86.2%, respectively, with accuracy of 86.7%.
What is known and what is new?
• PSA is widely used but has limitations in specificity and accuracy.
• This study introduced urinary GSTP1 methylation as a superior non-invasive diagnostic tool for prostate cancer (PCa), particularly in the PSA gray zone.
• The manuscript provided empirical evidence supporting the clinical utility of urinary GSTP1 methylation, outperforming PSA in specificity and accuracy.
• It introduced a diagnostic prediction model integrating GSTP1 methylation and PSA, significantly improving diagnostic efficacy.
What is the implication, and what should change now?
• The findings suggested that urinary GSTP1 methylation could be a valuable addition to current PCa diagnostic protocols, especially in challenging cases within the PSA gray zone. Future research should consider integrating urinary GSTP1 methylation testing to enhance PCa diagnosis, particularly for patients with PSA levels in the 4.0–10.0 ng/mL range.
Introduction
Globally, prostate cancer (PCa) representing the most prevalent malignancy in male. According to the global cancer data published by the World Health Organization in 2024, there are approximately 1,467,000 new cases and 397,000 deaths from PCa each year (1,2). In the United States, PCa is the second most common cause of cancer-related death in men (3). Early-stage PCa patients often lack noticeable symptoms, frequently diagnosed at intermediate or advanced stages, with rapid progression to metastatic castration-resistant PCa in some, marked by high invasiveness, incurability, and poor prognosis (4). Early diagnosis and intervention are crucial for improving survival rates and quality of life. Prostate biopsy is the gold standard for PCa diagnosis, and prostate-specific antigen (PSA) is the most widely used biomarker (5,6). However, the positive predictive value (PPV) of PSA as a screening tool is relatively low, and its low specificity frequently leads to unnecessary biopsies and overtreatment (7). Thus, the urgent need for a novel PCa biomarker with enhanced diagnostic specificity and sensitivity is evident, as it holds substantial clinical significance.
Researchers are increasingly focusing on DNA methylation, recognizing its role in various biological processes and disease mechanisms (8-10). DNA methylation is one of the earliest alterations in tumorigenesis (11), with genes like glutathione S-transferase Pi-1 (GSTP1) involved in cell proliferation, migration, and invasion (12,13). Studies on PCa tissue specimens show GSTP1 methylation detectable in early stages and grades, suggesting its role as an early carcinogenic event (14,15). It has been suggested as a diagnostic biomarker for assessing PCa biopsy samples, with the objective of minimizing the need for repetitive biopsies (16,17). GSTP1 methylation can be detected not only in PCa tissues and blood, but also in urine following prostate massage, with elevated methylation levels observed in the urine samples (18). Thus, the non-invasive urinary GSTP1 methylation testing technique has garnered significant interest for its potential application in PCa diagnosis. This study retrospectively analyzes clinical data from 120 suspected PCa patients who underwent prostate biopsy between September 2022 and June 2024 at The University of Hong Kong-Shenzhen Hospital, aiming to explore the diagnostic value of non-invasive urinary GSTP1 methylation testing for PCa. We present this article in accordance with the STARD reporting checklist (available at https://tau.amegroups.com/article/view/10.21037/tau-2024-689/rc).
Methods
General information
The study was conducted in accordance with the Declaration of Helsinki and its subsequent amendments. The study was approved by the Ethics Committee of The University of Hong Kong-Shenzhen Hospital (No. [2022]190) and individual consent for this retrospective analysis was waived due to the retrospective nature. This study encompassed patients admitted to the division of urology, department of surgery, The University of Hong Kong-Shenzhen Hospital from September 2022 to June 2024. Post clinical evaluation, all underwent transperineal prostate biopsy. Pre-biopsy, urine samples post-digital rectal examination (DRE) were collected for non-invasive urinary GSTP1 methylation testing. A total of 120 patients were included (Table 1), of which 59 were diagnosed with PCa. The remaining 61 were diagnosed with benign prostatic hyperplasia (BPH). All 120 patients underwent non-invasive GSTP1 methylation testing, with 58 predicted positive (i.e., PCa considered), including 48 in postoperative pathologically confirmed PCa patients, yielding a PPV of 82.8%.
Table 1
| Characteristics | BPH | PCa | P |
|---|---|---|---|
| Number of patients | 61 | 59 | |
| Age (years) | 67.9±6.3 | 70.8±8.8 | 0.043 |
| BMI (kg/m2) | 22.88±3.18 | 24.23±4.42 | 0.08 |
| Prostate volume (P1/4–3/4) (mL) | 64.0 (42.7, 88.6) | 40.1 (30.6, 61.3) | 0.001 |
| PSA value (P1/4–3/4) (ng/mL) | 9.6 (7.1, 15.1) | 18.0 (8.7, 53.9) | 0.002 |
| PI-RADS | |||
| 1 | 1 (1.6) | 0 (0.0) | <0.001 |
| 2 | 6 (9.7) | 0 (0.0) | |
| 3 | 26 (41.9) | 2 (3.4) | |
| 4 | 21 (34.4) | 22 (37.2) | |
| 5 | 7 (11.3) | 35 (60.3) |
Data are presented as number, mean ± standard deviation, n (%) or median (interquartile range). BMI, body mass index; BPH, benign prostatic hyperplasia; PCa, prostate cancer; PSA, prostate-specific antigen; PI-RADS, Prostate Imaging-Reporting and Data System.
Inclusion criteria
(I) Abnormal PSA levels [PSA >10.0 ng/mL; or 4.0–10.0 ng/mL with the ratio of free to total PSA (f/tPSA) <16%; PSA density (PSAD) >0.15 ng/mL2; annual PSA velocity >0.75 µg/L]; (II) pelvic magnetic resonance imaging (MRI), Prostate Imaging-Reporting and Data System (PI-RADS) ≥3; (III) prostate nodules on DRE; (IV) hypoechoic lesions identified on prostate ultrasound.
Exclusion criteria
(I) Acute infections (prostatitis/cystitis); (II) indwelling catheters; (III) severe coagulopathy/cardiopulmonary dysfunction; (IV) refusal to participate.
Withdrawal criteria
(I) Missing records/lost to follow-up; (II) substandard samples.
Method for detecting GSTP1 methylation in urine
Urine sample collection
Prior to prostate biopsy, a DRE is performed on the patient. After a 20-second massage during the DRE, the first 5–10 mL of urine is collected into a urine collection tube containing a preservative solution (Guangdong Huijin Chuangxing Biomedical Technology Co., Ltd., Guangzhou, China). The tube should be gently mixed to ensure thorough integration of the preservative with the urine. Samples can be stored at −20 ℃ for up to three months.
Urinary GSTP1 methylation detection
A total of 3.5 mL of urine was collected without centrifugation. Nucleic acid extraction and bisulfite conversion were automated using the AutoPure-12 instrument along with corresponding DNA extraction and bisulfite conversion reagents (all provided by Huijin Chuangxing Company). This process took approximately 3 hours, resulting in 100 µL of bisulfite-treated DNA (bisDNA). Subsequently, 10 µL of bisDNA was used as the template for quantitative polymerase chain reaction (qPCR). The GSTP1 methylation qPCR kit produced by Huijin Chuangxing Company was employed for the detection, following the manufacturer’s instructions. Each specimen underwent three technical replicates of qPCR. Upon completion of the qPCR reaction, each sample yielded two Ct values. The Ct value of the reference gene ACTB from the ROX channel was used to assess sample quality, and the Ct value of the GSTP1 from the FAM channel was utilized to determine the presence of methylated GSTP1 nucleic acid fragments. The relative methylation content of the target gene compared to the reference gene was calculated using the following formula (%): 2^(Ct ACTB − Ct Target) × 100 (19).
Result determination
(I) Results can be assessed when the ACTB Ct value is ≤36; otherwise, the sample testing result is considered invalid. (II) A GSTP1 Ct value of ≤41 indicates positive GSTP1methylation. (III) A GSTP1 Ct value of >41 indicates negative GSTP1 methylation. (IV) When ≥2 out of 3 replicates show positive methylation, the urine specimen is classified as methylation-positive.
Statistical analysis
Statistical analyses using SPSS v25.0 (IBM SPSS Inc., Armonk, NY, USA) evaluated patient data. Basic characteristics were analyzed via independent sample t-tests, χ2 tests, or rank sum tests. Kappa coefficient assessed consistency between diagnostic methods and the gold standard. Sensitivity, specificity, PPV, and accuracy for GSTP1 methylation and PSA in PCa-positive/BPH-negative biopsies were calculated. The area under the curve (AUC) were determined via receiver operator characteristic (ROC) curves, comparing diagnostic efficacy. Logistic regression assessed combined GSTP1 and PSA predictive probability. ROC curves and AUC evaluated combined method efficacy. P<0.05 was significant.
Results
Descriptive statistics of the patients
In this study, 120 patients who underwent prostate biopsy at The University of Hong Kong-Shenzhen Hospital from September 2022 to June 2024 were included (Figure 1). Among these, 59 patients were diagnosed with PCa, and the remaining 61 patients were diagnosed with BPH. Baseline characteristics of the included population are described in Table 1.
PCa diagnostic value of PSA and GSTP1 methylation
We compared the diagnostic efficacy of urine GSTP1 methylation and PSA. Urine GSTP1 methylation exhibited sensitivity of 81.4% and specificity of 83.6% for diagnosing PCa (Table 2), with a PPV of 82.8% and overall accuracy of 82.5%. In contrast, PSA had sensitivity of 67.8% and specificity of 54.1% (Table 3), with a PPV of 58.8% and overall accuracy of 60.8% (Table 4). Urine GSTP1 methylation outperformed PSA in sensitivity, specificity, and accuracy, showing strong diagnostic consistency (kappa =0.616) (Table 4). Urinary GSTP1 methylation showed no significant difference in sensitivity compared to PSA (P=0.16) (Table 5) but significantly higher specificity (P=0.001) (Table 6).
Table 2
| GSTP1 methylation | Prostate biopsy | Total | |
|---|---|---|---|
| + | - | ||
| + | 48 | 10 | 58 |
| - | 11 | 51 | 62 |
| Total | 59 | 61 | 120 |
Sensitivity =48/59×100%=81.4%; specificity =51/61×100%=83.6%. GSTP1, glutathione S-transferase Pi-1; PCa, prostate cancer.
Table 3
| PSA | Prostate biopsy | Total | |
|---|---|---|---|
| + | - | ||
| + | 40 | 28 | 68 |
| - | 19 | 33 | 52 |
| Total | 59 | 61 | 120 |
Sensitivity =40/59×100%=67.8%; specificity=33/61×100%=54.1%. PCa, prostate cancer; PSA, prostate-specific antigen.
Table 4
| Variables | Accuracy (%) | PPV (%) | NPV (%) | FPR (%) | FNR (%) | Kappa |
|---|---|---|---|---|---|---|
| PSA | 60.8 | 58.8 | 63.5 | 45.9 | 32.2 | 0.204 |
| GSTP1 methylation | 82.5 | 82.8 | 82.3 | 16.4 | 18.6 | 0.616 |
FNR, false negative rate; FPR, false positive rate; GSTP1, glutathione S-transferase Pi-1; NPV, negative predictive value; PCa, prostate cancer; PPV, positive predictive value; PSA, prostate-specific antigen.
Table 5
| PSA | GSTP1 methylation | Total | |
|---|---|---|---|
| + | - | ||
| + | 31 | 9 | 40 |
| - | 17 | 2 | 19 |
| Total | 48 | 11 | 59 |
P=0.16. GSTP1, glutathione S-transferase Pi-1; PCa, prostate cancer; PSA, prostate-specific antigen.
Table 6
| PSA | GSTP1 methylation | Total | |
|---|---|---|---|
| + | - | ||
| + | 4 | 24 | 28 |
| - | 6 | 27 | 33 |
| Total | 10 | 51 | 61 |
P=0.001. GSTP1, glutathione S-transferase Pi-1; PSA, prostate-specific antigen; PCa, prostate cancer.
The diagnostic efficacy of urinary GSTP1 methylation and PSA
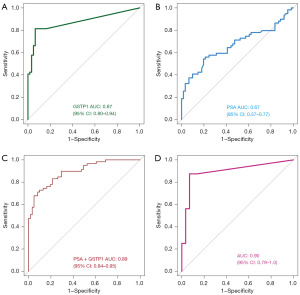
ROC curves were plotted to analyze diagnostic efficacy of urinary GSTP1 methylation and PSA. The AUC of the ROC for urinary GSTP1 methylation was 0.87 [95% confidence interval (CI): 0.80–0.94] (Figure 2A), and for PSA was 0.67 (95% CI: 0.57–0.77) (P<0.001) (Figure 2B), indicating superior diagnostic efficacy of GSTP1 methylation. Logistic regression assessed combined diagnostic value of GSTP1 and PSA in suspected PCa. The AUC of combined GSTP1 methylation and PSA was 0.89 (95% CI: 0.84–0.95), showing significant improvement over PSA alone (Figure 2C). In addition, the AUC of GSTP1 methylation in PSA gray zone was 0.90 (95% CI: 0.79–1.0) (Figure 2D).
Discussion
The incidence of PCa is escalating worldwide, particularly in Western countries, where it has emerged as the most prevalent malignancy among men (20). In recent years, China has also witnessed a significant surge in PCa cases, with a substantial proportion of patients being diagnosed at advanced stages (21). Therefore, early screening and diagnosis of PCa are critically important. PSA screening remains the cornerstone for the early detection of PCa. However, its low specificity frequently leads to unwarranted biopsies and overtreatment (6,22). In the resent years, GSTP1 methylation has garnered attention for its potential clinical relevance in early PCa detection, given that approximately 30% of PCa patients exhibit negative results on initial biopsy. Thus, GSTP1 has been suggested as a biomarker capable of mitigating the need for unnecessary re-biopsies (23). Ye et al. performed a meta-analysis on the PCa diagnostic efficacy of GSTP1 methylation in plasma (24), which revealed that the sensitivity and specificity of plasma GSTP1 methylation for diagnosing PCa were 37% and 97%, respectively, with an AUC of 0.78 (95% CI: 0.75–0.82). It is suggested that GSTP1 methylation in plasma exhibited high diagnostic specificity and could serve as a complementary diagnostic method to PSA. In addition, Wu et al. also reported that the diagnostic efficacy of GSTP1 methylation in plasma, serum, and prostate fluid samples for PCa diagnosis (25). The elevated methylation level of the GSTP1 gene was also detected in the urine of PCa patients (26). Urine has been identified as a practical medium for the non-invasive detection of PCa, given its accessibility (27). The quantification of one or multiple PCa-specific biomarkers in urine specimens has been employed for PCa diagnosis, offering a more convenient and non-invasive alternative to traditional liquid biopsy methods (28,29). Goessl and colleagues investigated the expression of GSTP1 gene methylation in urine samples following prostatic massage in patients with PCa, prostatic intraepithelial neoplasia (PIN), and BPH. The results demonstrated that GSTP1 promoter hypermethylation was present in 29 of 40 patients with PCa, in 2 of 7 patients with PIN, and in 1 of 42 patients with BPH (30). In Woodson et al.’s study, urine-based GSTP1 gene methylation detection demonstrated a specificity of 88% and a sensitivity of 91% for PCa. Additionally, their research confirmed a high correlation between GSTP1 gene methylation in urine and the methylation expression in biopsy tumor specimens (18). The aforementioned studies demonstrated the robust diagnostic capability of urine-based GSTP1 gene methylation. However, these studies on urine GSTP1 methylation typically used large-volume urine samples, which were centrifuged to obtain urine sediment. The technical steps involved in using large-volume urine samples made it challenging to automate methylation testing. We have improved this by developing a small-volume automated method for detecting GSTP1 methylation in urine, increasing detection efficiency and simplifying the process. In our study, we employed an automated method for detecting GSTP1 methylation in small-volume urine samples from patients with clinically suspected PCa. Our inclusion criteria were more stringent, we also compared the diagnostic performance of GSTP1 methylation with that of PSA. The results indicated that GSTP1 methylation had better diagnostic efficacy, with a diagnostic sensitivity and specificity of 81.4% and 83.6%, respectively, a PPV of 82.8%, a diagnostic accuracy of 82.5%, and a kappa value of 0.616. Its diagnostic sensitivity, specificity, and accuracy were superior to those of PSA, and the ROC curve had a larger AUC than that of PSA, resulting in better diagnostic efficacy. This method is non-invasive, easy to perform, and demonstrates high diagnostic accuracy. The use of our automated small-volume urine detection method for GSTP1 methylation can enhance the diagnostic performance for PCa, improve clinical diagnostic efficiency, and reduce the burden on patients. The mRNA quantification of PCa antigen 3 (PCA3) is an excellent urinary biomarker for PCa and was approved by the Food and Drug Administration (FDA) in the USA in 2012 for PCa screening (31). A meta-analysis of 46 clinical trials including 12,265 men who underwent PCA3 testing showed that the sensitivity and specificity for the detection of PCa were 65% and 73%, respectively (32). In another study that included 1,072 patients, the diagnostic AUC of PCA3 was 0.753 (33). Based on the above data, the diagnostic performance of urine GSTP1 gene methylation is comparable to that of PCA3, demonstrating its significant diagnostic potential. In the future, larger sample size studies will further validate the promising prospects of urine-based GSTP1 gene methylation.
In order to improve the diagnostic efficacy for PCa, researchers have explored the combined use of GSTP1 with other biomarkers. Bastian et al. reported that the combination of GSTP1 and several other DNA methylation markers, including DR1, EDNRB, NEP, RASSF1A and RAR-beta, may be useful biomarkers in men with hormone refractory PCa (34). Additionally, it is reported that noninvasive urine DNA methylation tests in combination with PSAD allowed the identification of clinically significant PCa (csPCa) with ≥90% sensitivity and specificity, which suggested that noninvasive urine DNA methylation tests in combination with PSAD could be used for further follow-up of the PCa patients (35). In our study, the results showed that the combined diagnostic specificity of GSTP1 methylation and PSA was 0.89 (95% CI: 0.80–0.95), which indicated that the combined diagnosis of urinary GSTP1 methylation and PSA in populations with suspected PCa demonstrates markedly superior diagnostic performance compared to PSA alone, leading to a significant increase in specificity and accuracy. Urinary GSTP1 methylation can be used as a complementary diagnostic method to compensate for the lack of specificity of PSA screening.
In the PSA “gray zone” (4.0–10.0 ng/mL), Stan et al. assessed plasma GSTP1 methylation’s diagnostic efficacy for PCa, retrospectively analyzing 80 patients with PSA levels between 4.0 and 10.0 ng/mL (36). The study found that for patients with PSA levels in this range, the sensitivity of plasma GSTP1 methylation for diagnosing PCa was 69.81%, with a specificity of 81.48% and a PPV of 88.1%. This indicated that plasma GSTP1 methylation have significant diagnostic efficacy in suspected PCa patients. In addition, another study reported that within the PSA range of 2.0 to 10.0 ng/mL, the sensitivity of plasma GSTP1 methylation ranged from 58.1% to 69.4%, with specificity between 76.7% and 81.6%, and predictive accuracy between 72% and 77%, yielding an AUC of 0.68 in ROC curve analysis (37). Therefore, in our study, we explored the diagnostic efficacy of urinary GSTP1 methylation in the PSA “gray zone”. The results demonstrated that the sensitivity, specificity, and accuracy of urinary GSTP1 methylation in the PSA “gray zone” were 87.5%, 86.2%, and 86.7%, respectively. The AUC for urinary GSTP1 methylation in the PSA “gray zone” population was 0.90 (95% CI: 0.79–1.0). Compared to plasma GSTP1 methylation, urinary GSTP1 methylation exhibited similar diagnostic performance and holds potential research value. However, due to the limited sample size, these findings require validation in larger studies.
Conclusions
Urinary GSTP1 methylation demonstrated promising diagnostic efficacy in the detection of suspected PCa patients, with the potential to serve as a biomarker for PCa. The combination of urinary GSTP1 methylation and PSA can enhance diagnostic specificity and accuracy. Urinary GSTP1 methylation showed good diagnostic performance when PSA levels were between 4.0–10.0 ng/mL, offering significant research value. Additionally, the use of small-volume urine samples for automated GSTP1 methylation testing offers advantages such as non-invasive collection, ease of detection, and high diagnostic efficacy, indicating strong potential for clinical application. However, as this study was a single-center retrospective analysis, further validation through multi-center, prospective, and larger-scale studies are required to confirm these findings and assess its clinical value for PCa screening and diagnosis.
Acknowledgments
None.
Footnote
Reporting Checklist: The authors have completed the STARD reporting checklist. Available at https://tau.amegroups.com/article/view/10.21037/tau-2024-689/rc
Data Sharing Statement: Available at https://tau.amegroups.com/article/view/10.21037/tau-2024-689/dss
Peer Review File: Available at https://tau.amegroups.com/article/view/10.21037/tau-2024-689/prf
Funding: None.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tau.amegroups.com/article/view/10.21037/tau-2024-689/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki and its subsequent amendments. The study was approved by the Ethics Committee of The University of Hong Kong-Shenzhen Hospital (No. [2022]190) and individual consent for this retrospective analysis was waived due to the retrospective nature.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Laversanne M, Sung H, et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2024;74:229-63. [Crossref] [PubMed]
- Heimdörfer D, Artamonova N, Culig Z, et al. Unraveling molecular characteristics and tumor microenvironment dynamics of neuroendocrine prostate cancer. J Cancer Res Clin Oncol 2024;150:462. [Crossref] [PubMed]
- Siegel RL, Kratzer TB, Giaquinto AN, et al. Cancer statistics, 2025. CA Cancer J Clin 2025;75:10-45. [Crossref] [PubMed]
- Zhou Y, Ou L, Xu J, et al. FAM64A is an androgen receptor-regulated feedback tumor promoter in prostate cancer. Cell Death Dis 2021;12:668. [Crossref] [PubMed]
- Emmett L, Papa N, Counter W, et al. Reproducibility and Accuracy of the PRIMARY Score on PSMA PET and of PI-RADS on Multiparametric MRI for Prostate Cancer Diagnosis Within a Real-World Database. J Nucl Med 2024;65:94-9. [Crossref] [PubMed]
- Brikun I, Nusskern D, Decatus A, et al. A panel of DNA methylation markers for the detection of prostate cancer from FV and DRE urine DNA. Clin Epigenetics 2018;10:91. [Crossref] [PubMed]
- Perera M, Manning T, Finelli A, et al. Management of men with previous negative prostate biopsy. Curr Opin Urol 2016;26:481-7. [Crossref] [PubMed]
- Naik A, Thakur N. Epigenetic regulation of TGF-β and vice versa in cancers - A review on recent developments. Biochim Biophys Acta Rev Cancer 2024;1879:189219. [Crossref] [PubMed]
- Chen Y, Dong GH, Li S, et al. The associations between exposure to ambient air pollution and coagulation markers and the potential effects of DNA methylation. J Hazard Mater 2024;480:136433. [Crossref] [PubMed]
- Witzig TE, Taylor WR, Mahoney DW, et al. Blood Plasma Methylated DNA Markers in the Detection of Lymphoma: Discovery, Validation, and Clinical Pilot. Am J Hematol 2025;100:218-28. [Crossref] [PubMed]
- Liu Y, Zhang Y, Du D, et al. PCDH17 is regulated by methylation of DNMT3B and affects the malignant biological behavior of HCC through EMT. Exp Cell Res 2022;418:113245. [Crossref] [PubMed]
- Ye S, Ni Y. lncRNA SNHG9 Promotes Cell Proliferation, Migration, and Invasion in Human Hepatocellular Carcinoma Cells by Increasing GSTP1 Methylation, as Revealed by CRISPR-dCas9. Front Mol Biosci 2021;8:649976. [Crossref] [PubMed]
- Li N, Zhao Z, Miao F, et al. Silencing of long non-coding RNA LINC01270 inhibits esophageal cancer progression and enhances chemosensitivity to 5-fluorouracil by mediating GSTP1methylation. Cancer Gene Ther 2021;28:471-85. [Crossref] [PubMed]
- Zelic R, Fiano V, Zugna D, et al. Global Hypomethylation (LINE-1) and Gene-Specific Hypermethylation (GSTP1) on Initial Negative Prostate Biopsy as Markers of Prostate Cancer on a Rebiopsy. Clin Cancer Res 2016;22:984-92. [Crossref] [PubMed]
- Aykanli E, Arisan S, Arisan ED, et al. Diagnostic Value of GSTP1, RASSF1, AND RASSF2 Methylation in Serum of Prostate Cancer Patients. Urol J 2024;21:182-8.
- Stewart GD, Van Neste L, Delvenne P, et al. Clinical utility of an epigenetic assay to detect occult prostate cancer in histopathologically negative biopsies: results of the MATLOC study. J Urol 2013;189:1110-6. [Crossref] [PubMed]
- Mahon KL, Qu W, Lin HM, et al. Serum Free Methylated Glutathione S-transferase 1 DNA Levels, Survival, and Response to Docetaxel in Metastatic, Castration-resistant Prostate Cancer: Post Hoc Analyses of Data from a Phase 3 Trial. Eur Urol 2019;76:306-12. [Crossref] [PubMed]
- Woodson K, O'Reilly KJ, Hanson JC, et al. The usefulness of the detection of GSTP1 methylation in urine as a biomarker in the diagnosis of prostate cancer. J Urol 2008;179:508-11; discussion 511-2. [Crossref] [PubMed]
- De Strooper LM, Meijer CJ, Berkhof J, et al. Methylation analysis of the FAM19A4 gene in cervical scrapes is highly efficient in detecting cervical carcinomas and advanced CIN2/3 lesions. Cancer Prev Res (Phila) 2014;7:1251-7. [Crossref] [PubMed]
- Siegel RL, Giaquinto AN, Jemal A. Cancer statistics, 2024. CA Cancer J Clin 2024;74:12-49. [Crossref] [PubMed]
- Cao W, Chen HD, Yu YW, et al. Changing profiles of cancer burden worldwide and in China: a secondary analysis of the global cancer statistics 2020. Chin Med J (Engl) 2021;134:783-91. [Crossref] [PubMed]
- Xiang J, Yan H, Li J, et al. Transperineal versus transrectal prostate biopsy in the diagnosis of prostate cancer: a systematic review and meta-analysis. World J Surg Oncol 2019;17:31. [Crossref] [PubMed]
- Van Neste L, Herman JG, Otto G, et al. The epigenetic promise for prostate cancer diagnosis. Prostate 2012;72:1248-61. [Crossref] [PubMed]
- Ye J, Wu M, He L, et al. Glutathione-S-Transferase p1 Gene Promoter Methylation in Cell-Free DNA as a Diagnostic and Prognostic Tool for Prostate Cancer: A Systematic Review and Meta-Analysis. Int J Endocrinol 2023;2023:7279243. [Crossref] [PubMed]
- Wu T, Giovannucci E, Welge J, et al. Measurement of GSTP1 promoter methylation in body fluids may complement PSA screening: a meta-analysis. Br J Cancer 2011;105:65-73. [Crossref] [PubMed]
- Nakayama M, Gonzalgo ML, Yegnasubramanian S, et al. GSTP1 CpG island hypermethylation as a molecular biomarker for prostate cancer. J Cell Biochem 2004;91:540-52. [Crossref] [PubMed]
- Guo J, Liu D, Zhang X, et al. Establishing a Urine-Based Biomarker Assay for Prostate Cancer Risk Stratification. Front Cell Dev Biol 2020;8:597961. [Crossref] [PubMed]
- Groskopf J, Aubin SM, Deras IL, et al. APTIMA PCA3 molecular urine test: development of a method to aid in the diagnosis of prostate cancer. Clin Chem 2006;52:1089-95. [Crossref] [PubMed]
- Guo J, Yang J, Zhang X, et al. A Panel of Biomarkers for Diagnosis of Prostate Cancer Using Urine Samples. Anticancer Res 2018;38:1471-7. [Crossref] [PubMed]
- Goessl C, Müller M, Heicappell R, et al. DNA-based detection of prostate cancer in urine after prostatic massage. Urology 2001;58:335-8. [Crossref] [PubMed]
- Fujita K, Nonomura N. Urinary biomarkers of prostate cancer. Int J Urol 2018;25:770-9. [Crossref] [PubMed]
- Cui Y, Cao W, Li Q, et al. Evaluation of prostate cancer antigen 3 for detecting prostate cancer: a systematic review and meta-analysis. Sci Rep 2016;6:25776. [Crossref] [PubMed]
- Aubin SM, Reid J, Sarno MJ, et al. PCA3 molecular urine test for predicting repeat prostate biopsy outcome in populations at risk: validation in the placebo arm of the dutasteride REDUCE trial. J Urol 2010;184:1947-52. [Crossref] [PubMed]
- Bastian PJ, Palapattu GS, Yegnasubramanian S, et al. CpG island hypermethylation profile in the serum of men with clinically localized and hormone refractory metastatic prostate cancer. J Urol 2008;179:529-34; discussion 534-5. [Crossref] [PubMed]
- Matulevičius A, Žukauskaitė K, Gineikaitė R, et al. Combination of DNA methylation biomarkers with multiparametric magnetic resonance and ultrasound imaging fusion biopsy to detect the local spread of prostate cancer. Prostate 2023;83:1572-83. [Crossref] [PubMed]
- Stan M, Botnarciuc V, Suceveanu AI, et al. Role of glutathione-S-transferase gene P1 in the diagnosis of prostate cancer in patients with 'grey level' prostate-specific antigen values. Exp Ther Med 2022;24:591. [Crossref] [PubMed]
- Baden J, Adams S, Astacio T, et al. Predicting prostate biopsy result in men with prostate specific antigen 2.0 to 10.0 ng/ml using an investigational prostate cancer methylation assay. J Urol 2011;186:2101-6. [Crossref] [PubMed]


