
PRRX1/FOXM1 reduces gemcitabin-induced cytotoxicity by regulating autophagy in bladder cancer
Introduction
Bladder cancer (BC) is among the most common malignancies affecting the urinary system, the morbidity and mortality of which have risen in recent years (1,2). Although there has been considerable effort and progress in BC treatment, knowledge of the mechanisms of BC tumorigenesis and progression is still lacking, and the survival rate of patients with BC remains unsatisfactory (3-5). Therefore, it is of great significance to further explore important regulatory mechanisms in BC and to look for sensitive biomarkers for the treatment of this disease.
Increasing studies have identified that proteins exert marked regulatory effects on cell biological processes to affect the progression of various diseases (6-8). Paired related homeobox 1 (PRRX1), the paired-related homeobox transcription factor, induces epithelial-mesenchymal transition (EMT) in tumors, leading to a poor prognosis (9-12). In salivary adenoid cystic carcinoma, PRRX1-induced EMT stimulates the metabolic reprogramming of free fatty acids to facilitate tumor invasion and metastasis (13). Transcription factor PRRX1 accelerates the differentiation of brown adipose-derived stem cells into sinus node-like cells (14). PRRX1 also activates the TGF-β/Smad pathway to aggravate stemness and angiogenesis in glioma (15), and modulates cellular phenotype plasticity and dormancy via miR-642b-3p in head and neck squamous cell carcinoma (16). However, the role of PRRX1 in the progression of BC remains unclear. Therefore, the focus of the present study was to explore the role and related mechanisms of PRRX1 in BC.
Gemcitabine serves as the first-line cytotoxic chemotherapeutic drug for BC (17,18). However, although gemcitabine treatment has some clinical benefits, it offers only limited control over the progression of BC. Gemcitabine initially works well for pancreatic, breast, bladder, and non-small cell lung cancers, but after several rounds of treatment, most patients gradually become unresponsive, leading to tumor recurrence (19-21). Therefore, gaining an in-depth understanding of the molecular mechanisms related to gemcitabine-induced cytotoxicity is important, and searching for novel drug markers is a feasible strategy to reverse drug resistance. Recent studies have revealed that suppressing Src decreases gemcitabine-induced cytotoxicity in human pancreatic cancer cell lines (22), and that everolimus strengthens gemcitabine-induced cytotoxicity in BC cell lines (23). Furthermore, in mantle cell lymphoma, silencing of HDM-2 has been found to repress the expression of ribonucleotide reductase subunit M2 while synergistically strengthening gemcitabine-induced cytotoxicity (24). However, the relationship between gemcitabine-induced cytotoxicity and PRRX1 in BC is unclear.
Autophagy is a vital process to participate into cancers’ progression, also in BC. For example, lncRNA ADAMTS9-AS1 regulates PI3K/AKT/mTOR signaling pathway to suppress cell apoptosis and autophagy in BC (25). In addition, artesunate enhances ROS and activates AMPK-mTOR-ULK1 axis to stimulate autophagy dependent apoptosis in BC (26). Besides, leflunomide retards autophagy and the PI3K/Akt signaling pathway to suppress BC tumorigenesis (27). But, the relevant regulatory roles of PRRX1 on autophagy in BC need further investigations.
In the present study, we aimed to shed light on the role of PRRX1 and its effects on gemcitabine-induced cytotoxicity in BC. Our study found that PRRX1 affected gemcitabine-induced cytotoxicity in BC cells by regulating the expression of the autophagy proteins LC3 and Beclin-1. This observation suggests that PRRX1 might serve as a novel biomarker of gemcitabine-induced cytotoxicity in patients receiving BC treatment. We present the following article in accordance with the ARRIVE reporting checklist (available at https://tau.amegroups.com/article/view/10.21037/tau-22-415/rc).
Methods
Tissue samples
Sample BC tissues (n=75) and adjacent normal tissues (n=75) were collected from patients receiving treatment for BC at The First Affiliated Hospital of Wenzhou Medical University. After collection, the tissues were immediately stored at −80 ℃.
Other samples were offered from the urology sample bank of The First Affiliated Hospital of Wenzhou Medical University. Approval for this study was obtained from the Ethics Committee of The First Affiliated Hospital of Wenzhou Medical University (Approval. No. 2017-097). The study was in accordance with the Helsinki Declaration (as revised in 2013). Informed consent was taken from all the patients.
Cell culture
Five BC cell lines (T24, RT4, J82, SW780, and 5637) and the normal human bladder epithelial cell line HCV-29 were purchased from the American Type Culture Collection (ATCC; Manassas, VA, USA). Dulbecco’s Modified Eagle Medium (DMEM, Gibco, USA) supplemented with 10% fetal bovine serum (FBS) was applied to cultivate the cells, which were then incubated at 37 ℃ under a humidified atmosphere with 5% CO2.
Cell transfection
Short hairpin RNA (shRNA) targeting PRRX1 (sh1#PRRX1 and sh2#PRRX1) and FOXM1 (sh1#FOXM1 and sh2#FOXM1) and negative control (shNC), along with pcDNA3.1/PRRX1 and pcDNA3.1/FOXM1 with negative control (empty pcDNA3.1), were purchased from GenePharma (Shanghai, China). Transfection of the above vectors into BC cells was conducted using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA).
Western blot
Proteins were extracted from BC tissue samples and cells using RIPA reagent, and quantified using a bicinchoninic acid (BCA) protein quantitation kit. Then, sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) was performed for protein separation, after which the proteins were moved onto polyvinylidene fluoride membranes. After being sealed with 5% defatted milk, the membranes were incubated with primary antibodies and then with horseradish peroxidase-conjugated secondary antibody. The following antibodies were used, all of which were bought from Abcam (Shanghai, China): PRRX1 (1:1,000, ab106834; Abcam), LC3B (1:3,000, ab63817; Abcam, containing LC3-I and LC3-II), Beclin-1 (1:1,000, ab210498; Abcam), FOXM1 (1:1,000, ab245309; Abcam), and β-actin (1:1,000, ab8226; Abcam). An enhanced electrochemiluminescence (ECL) detection system (Millipore) was used to detect the protein bands.
In vivo investigation
Animal experiments were performed under a project license (Approval No. WYDW2021-0014) granted by the Ethics Committee of Wenzhou Medical University, in compliance with the National Institutes of Health Laboratory guidelines for the care and use of animals. Five-week-old male BALB/c nude mice were divided into four groups (n=8 in each group): the shNC, sh1#PRRX1, gemcitabine + shNC, and gemcitabine + sh1#PRRX1 groups. All mice were purchased from the Vital River company (Beijing, China). Transfected BC cells were subcutaneously inoculated into the right flank of each nude mouse. The tumor volumes were estimated every 7 days using the formula: Volume =1/2 (length × width2). After 4 weeks, the mice were sacrificed by cervical dislocation under deep anesthesia with 2% isoflurane (Baxter Healthcare Corporation, New Providence, NJ, USA), and their tumors were photographed and weighed.
Flow cytometry
A cell apoptosis assay was performed using the Annexin V-FITC Apoptosis Detection Kit (Sigma-Aldrich, St. Louis, MO, USA). Transfected BC cells were collected and subjected to two rinses with cold phosphate-buffered saline followed by incubation with Annexin V-FITC (fluorescein isothiocyanate) and propidium iodide at room temperature in the dark. Finally, cell apoptosis was examined through flow cytometry (BD Biosciences, San Jose, CA, USA).
Green fluorescent protein (GFP)-LC3 immunofluorescence assay
Autophagy was assessed through GFP-LC3 immunofluorescence analysis. The transfection of GFP-LC3 plasmids was performed using Lipofectamine 2000 reagent (Invitrogen). The LC3 protein was shown as the aggregated green fluorescent particles. Fluorescence images were acquired and analyzed using a fluorescence microscope (Nikon Corporation, Tokyo, Japan).
MTT assay
Cell viability and the half-maximal inhibitory concentration (IC50) were assessed by MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide) assay. Briefly, BC cells were plated in 96-well plates and MTT solution (10 µL, 5 mg/mL) was added into each well. After 4-hour incubation, the culture medium was removed, and dimethyl sulfoxide (100 µL) was added to dissolve the crystals. The absorbance was evaluated at 490 nm with a microplate reader (Bio-Tek Instruments, USA). Gemcitabine at different doses was applied to treat BC cells.
Sphere formation assay
The BC cells were planted on the 6-well plates (Corning, USA). The plates were kept in one incubator for 2 weeks with 5% CO2 at 37 ℃. Then, the formed colonies were made to fixing with 4% paraformaldehyde and staining with 0.1% crystal violet. The images were obtained under a microscope.
Real-time quantitative polymerase chain reaction (RT-qPCR) assay
Isolation of total RNA was conducted using TRIzol (Life technologies, USA) and then complementary DNA was reverse-transcribed using the PrimeScript RT Reagent Kit (TaKaRa, Dalian, China). The polymerase chain reaction (PCR) reactions were done using SYBR® Premix Ex Taq™ (TaKaRa, Dalian, China) on the ABI Prism 7500 Fast Real-Time PCR system (Applied Biosystems, USA). β-actin was employed as the internal control for genes. The 2-ΔΔCt method was used to calculate the expression levels of target genes. The PCR primers used were as follow:
PRRX1-F 5ʹ-CAGGCGGATGAGAACGTGG-3ʹ;
PRRX1-R 5ʹ-AAAAGCATCAGGATAGTGTGTCC-3ʹ;
FOXM1-F 5ʹ-ACTTTAAGCACATTGCCAAGC-3ʹ;
FOXM1-R 5ʹ-CGTGCAGGGAAAGGTTGT-3ʹ;
LC3B-F 5ʹ-GATGTCCGACTTATTCGAGAGC-3ʹ;
LC3B-R 5ʹ-TTGAGCTGTAAGCGCCTTCTA-3ʹ;
Beclin-1-F 5ʹ-GGATGGATGTGGAGAAAGGCAAG-3ʹ;
Beclin-1-R 5ʹ-TGAGGACACCCAAGCAAGACC-3ʹ;
β-actin-F 5ʹ-CTCCATCCTGGCCTCGCTGT-3ʹ;
β-actin-R 5ʹ-GCTGTCACCTTCACCGTTCC-3ʹ.
Immunohistochemical (IHC) staining assay
An IHC assay was performed with formalin-fixed, paraffin-embedded tissues. After slicing, the tissues were deparaffinized and rehydrated. The tissues were first incubated with anti-Ki-67, anti-PRRX1, and anti-Beclin-1 at 4 ℃ overnight, and then probed with secondary antibodies with high-sensitivity streptavidin–horseradish peroxidase conjugate. For observation, slides were dyed with 3, 3'-diaminobenzidine and counterstained with Hematoxylin-eosin staining.
Statistical analysis
Data were denoted as mean ± SD (standard deviation). Statistical analyses were conducted using the SPSS 20.0 software (Chicago, IL, USA). All experiments were done in triplicate. Differences among groups were analyzed using Student’s t-test (for two groups) or one-way ANOVA (for more than two groups). Statistical significance was indicated by P<0.05.
Results
PRRX1 was highly expressed in BC tissues
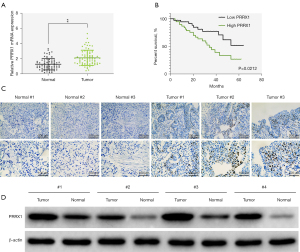
Abnormal expression of PRRX1 has been confirmed to affect the progression of a variety of tumors (13,14,16,28). The expression of PRRX1 in TCGA-BLCA database was analyzed by online tool GEPIA, and the results showed no significant difference of PRRX1 mRNA expression in normal tissues and BC tissues (Figure S1A). But, the prognosis of BC patients with high PRRX1 expression was significantly poor (Figure S1B). In this study, we first verified the expression of PRRX1 in BC tissues. Results of real-time quantitative PCR analysis revealed that PRRX1 expression was higher in BC tissues than in adjacent normal tissues (Figure 1A). Further, patients with BC who had high levels of PRRX1 had a notably poorer prognosis than those with low PRRX1 levels (Figure 1B). Through IHC and western blot analyses, PRRX1 expression was found to be up-regulated in the tumor group compared with the normal group (Figure 1C,1D). Together, these results showed that PRRX1 was highly expressed in BC tissues.
PRRX1 was up-regulated in BC cells and enhanced cell viability
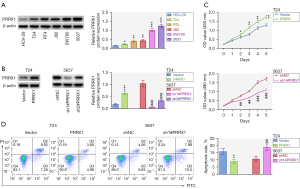
To explore the possible biological effect of PRRX1 on BC cell phenotype, we detected the protein expression of PRRX1 in the BC cell lines T24, RT4, J82, SW780 and 5637. Results showed that PRRX1 protein expression was higher in BC cell lines than in the human bladder epithelial cell line HCV-29 (Figure 2A). Confirmation of the overexpression and knockdown efficiency of PRRX1 is shown in Figure 2B. Cell viability was enhanced with PRRX1 up-regulation but reduced after PRRX1 was repressed (Figure 2C). Moreover, PRRX1 overexpression decreased the cell apoptosis rate, while PRRX1 suppression had the opposite effect (Figure 2D). These data indicated that PRRX1 was up-regulated in BC cells and accelerated cell viability.
PRRX1 weakened gemcitabine-induced cytotoxicity and enhanced gemcitabine-induced autophagy in BC cells
Many reports have evidenced that gemcitabine is an effective therapeutic agent for BC (23,29,30). Therefore, we next investigated the effects of PRRX1 on gemcitabine-induced cytotoxicity in BC cells. The gemcitabine reduced the cell viability of T24 and 5,637 cells with a dose-dependent manner (Figure S1C). Additionally, gemcitabine treatment (1 µM) decreased the cell proliferation abilities of T24 and 5,637 cells (Figure S1D). As shown in Figure 3A, PRRX1 overexpression strengthened the gemcitabine-induced decreases in cell viability and the IC50 in BC cells, whereas silencing of PRRX1 had the opposite effect. After gemcitabine treatment, cell apoptosis was enhanced; however, this effect could be offset by overexpressing PRRX1 or further aggravated by inhibiting PRRX1 (Figure 3B).
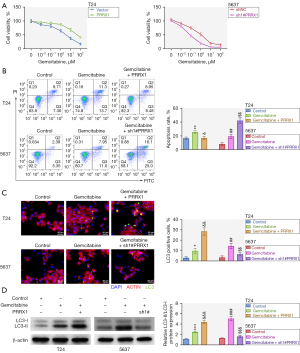
Next, we further studied the effects of PRRX1 on autophagy. The gemcitabine-induced enhancement of GFP-LC3 puncta formation and the number of GFP-LC3-positive cells were exacerbated by PRRX1 overexpression but reversed by PRRX1 knockdown (Figure 3C). Furthermore, gemcitabine treatment increased the expression of LC3II/I, and this effect could be enhanced by overexpressing PRRX1 and reversed by suppressing PRRX1 (Figure 3D). In summary, PRRX1 weakened gemcitabine-induced cytotoxicity and enhanced gemcitabine-induced autophagy in BC cells.
PRRX1 up-regulated the expression of LC3 and Beclin-1 through cooperation with FOXM1
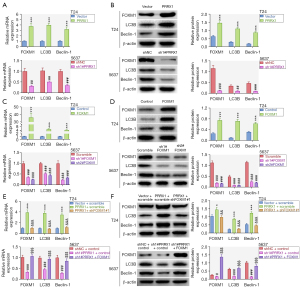
Our results showed that the mRNA and protein expression levels of FOXM1, LC3B, and Beclin-1 were increased by PRRX1 overexpression and decreased by PRRX1 silencing (Figure 4A,4B). Previous research has revealed that PRRX1 cooperates with FOXM1 to regulate downstream genes (31). FOXM1 has been discovered to play a role in autophagy through transcriptionally regulating the Beclin-1 and LC3 genes in human triple-negative breast cancer cells (32). In this study, overexpression of FOXM1 elevated, and knockdown of FOXM1 diminished, the mRNA and protein expression of FOXM1, LC3B, and Beclin-1 (Figure 4C,4D). The increases in LC3B and Beclin-1 mRNA and protein expression that were induced by PRRX1 overexpression could be reversed by silencing FOXM1. Also, the decreases in FOXM1, LC3B, and Beclin-1 mRNA and protein expression caused by PRRX1 inhibition could be counteracted by up-regulating FOXM1 (Figure 4E,4F). These findings indicated that PRRX1 up-regulated the expression of LC3 and Beclin-1 by cooperating with FOXM1.
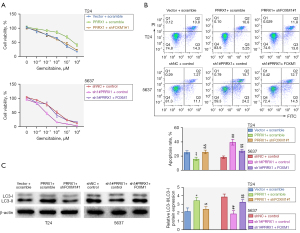
PRRX1 weakened gemcitabine-induced cytotoxicity and induced autophagy through FOXM1
Subsequently, we investigated whether PRRX1 weakened gemcitabine-induced cytotoxicity and induced autophagy through FOXM1. Cell viability and the IC50 were enhanced after PRRX1 overexpression, but silencing of FOXM1 reversed this effect. Meanwhile, cell viability and the IC50 were decreased after PRRX1 suppression, but FOXM1 overexpression rescued this effect (Figure 5A). The decrease in cell apoptosis induced by PRRX1 overexpression could be neutralized by repressing FOXM1, and the increase in cell apoptosis caused by PRRX1 silencing could be neutralized by overexpressing FOXM1 (Figure 5B). Furthermore, knockdown of FOXM1 reversed the enhancing effect that PRRX1 up-regulation had on LC3II/I expression, while overexpression of FOXM1 counteracted the reduction in LC3II/I expression caused by repressing PRRX1 (Figure 5C). In summary, PRRX1 weakened gemcitabine-induced cytotoxicity and induced autophagy by regulating FOXM1.
Knockdown of PRRX1 enhanced the inhibitive effects of gemcitabine on tumor growth in vivo
Finally, the role of PRRX1 in BC was investigated in vivo. The size, volume, and weight of mouse tumors were decreased with PRRX1 suppression and with gemcitabine treatment, and this gemcitabine-induced effect was further strengthened by PRRX1 knockdown (Figure 6A). Also, PRRX1 suppression deeply aggravated the decreases in PRRX1, Ki67, and Beclin-1 expression observed after PRRX1 silencing and after gemcitabine treatment (Figure 6B). These results revealed that PRRX1 knockdown enhanced the inhibitive effects of gemcitabine on tumor growth in vivo.

Discussion
Evidence shows that dysregulation of PRRX1 influences the progression of some tumors (10,13-16). However, the involvement of PRRX1 in BC progression has yet to be fully understood. In this work, PRRX1 exhibited higher expression in BC tissues and cells than that in normal bladder tissues and cells, with high PRRX1 expression being related to poor overall survival in patients with BC. Furthermore, PRRX1 was found to accelerate cell viability and hinder cell apoptosis. Our study also examined the relationship between PRRX1 and cytotoxicity induced by gemcitabine (the first-line chemotherapeutic drug for BC). Gemcitabine has obtained more and more attention for its cytotoxic effects in patients with BC (23,29,30); however, the relationship between gemcitabine-induced cytotoxicity and PRRX1 in BC has remained unclear. Our results showed that PRRX1 weakened gemcitabine-induced cytotoxicity in BC cells.
Autophagy is a cellular process in which misfolded proteins and malfunctional organelles target lysosomes or vacuoles for degradation (33,34). Autophagy has two roles: firstly, it can induce tumor cell death; secondly, it can suppress cell stimulation and play a supporting role in tumor cell survival (35,36). The effects of autophagy on BC have been investigated in many studies. For example, Wang et al. reported that chloroquine suppresses autophagy and induces apoptosis to heighten the radiosensitivity of BC cells (37). Kou et al. found that autophagy induction regulates the AMPK/mTOR pathway to increase tetrandrine-induced apoptosis in human BC cells (38). A study by Hung et al. uncovered the role of rhopaloic acid A in modulating ROS-mediated signaling to stimulate autophagy, apoptosis, and MAPK activation in BC (39). In their study, Fu et al. reported that hypoxia-induced autophagy activates hypoxia-inducible factor 1α in BC to facilitate gemcitabine resistance (40). In this work, we found that PRRX1 enhanced the GFP-LC3 puncta formation, the number of GFP-LC3-positive cells, and the expression of LC3II/I to induce autophagy in BC cells.
Many proteins cooperate with other proteins to regulate cell biological processes. For instance, HSPB8 cooperates with BAG3 to accelerate the spatial sequestration of ubiquitinated proteins and coordinate the adaptive response of cells to proteasome dysfunction (41). JIP1 and JIP3 cooperate to activate kinesin-1, which mediates TrkB anterograde axonal transport (42). DAPLE binds with MPDZ to accelerate apical cell constriction (43). The RNA binding protein RBMS3 interacts with PRRX1 to affect the progression of triple-negative breast cancer (44). PRRX1 cooperates with FOXM1 to regulate downstream genes (31), and FOXM1 has been revealed to play a role in autophagy by transcriptionally regulating the Beclin-1 and LC3 genes (32). Therefore, we speculated that PRRX1 cooperates with FOXM1 to regulate autophagy in BC. In this study, PRRX1 was verified to up-regulate the expression of LC3 and Beclin-1 by cooperating with FOXM1. Results from rescue assays demonstrated that FOXM1 reversed the effects of PRRX1 on gemcitabine-induced cytotoxicity and autophagy, while knockdown of PRRX1 enhanced the inhibitive effects of gemcitabine on tumor growth in vivo.
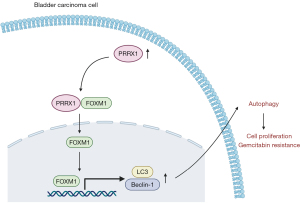
In summary, the results of this study suggest that PRRX1 cooperates with FOXM1 to reduce gemcitabine-induced cytotoxicity in BC cells by regulating the expression of the autophagy proteins LC3 and Beclin-1 (Figure 7). This discovery may offer a potential biological target for BC treatment. However, our study still had some limitations (no studies on other drugs, other cellular progresses, other BC animal model), and we will address these by performing more experiments for further exploration in the future. This study suggested that PRRX1/FOXM1 axis might be an effective bio-target for BC treatment and prognosis.
Acknowledgments
Funding: This work was supported by the Wenzhou Science and Technology Bureau (grant No. Y2020949).
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at https://tau.amegroups.com/article/view/10.21037/tau-22-415/rc
Data Sharing Statement: Available at https://tau.amegroups.com/article/view/10.21037/tau-22-415/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tau.amegroups.com/article/view/10.21037/tau-22-415/coif). All authors report funding support from the Wenzhou Science and Technology Bureau (grant No. Y2020949). The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. Approval for this study was obtained from the Ethics Committee of The First Affiliated Hospital of Wenzhou Medical University (Approval No. 2017-097). The study was in accordance with the Helsinki Declaration (as revised in 2013). Informed consent was taken from all the patients. Animal experiments were performed under a project license (Approval No. WYDW2021-0014) granted by the Ethics Committee of Wenzhou Medical University, in compliance with the National Institutes of Health Laboratory guidelines for the care and use of animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Chavan S, Bray F, Lortet-Tieulent J, et al. International variations in bladder cancer incidence and mortality. Eur Urol 2014;66:59-73. [Crossref] [PubMed]
- Dobruch J, Daneshmand S, Fisch M, et al. Gender and Bladder Cancer: A Collaborative Review of Etiology, Biology, and Outcomes. Eur Urol 2016;69:300-10. [Crossref] [PubMed]
- Alifrangis C, McGovern U, Freeman A, et al. Molecular and histopathology directed therapy for advanced bladder cancer. Nat Rev Urol 2019;16:465-83. [Crossref] [PubMed]
- Chou R, Selph SS, Buckley DI, et al. Treatment of muscle-invasive bladder cancer: A systematic review. Cancer 2016;122:842-51. [Crossref] [PubMed]
- Nativ O, Dalal E, Sabo E, et al. Halofuginone: a novel oral and intravesical agent for the treatment of non-muscle invasive bladder cancer. Journal of Molecular and Clinical Medicine 2018;1:213-7.
- Tahmasebi S, Khoutorsky A, Mathews MB, et al. Translation deregulation in human disease. Nat Rev Mol Cell Biol 2018;19:791-807. [Crossref] [PubMed]
- Zoghbi HY, Beaudet AL. Epigenetics and Human Disease. Cold Spring Harb Perspect Biol 2016;8:a019497. [Crossref] [PubMed]
- Ozer V, Karaca Y, Gunaydin M, et al. Serum Irisin Levels in Patients with Acute Atrial Fibrillation. Available online: https://www.signavitae.com/articles/10.22514/SV152.092019.5
- Takahashi Y, Sawada G, Kurashige J, et al. Paired related homoeobox 1, a new EMT inducer, is involved in metastasis and poor prognosis in colorectal cancer. Br J Cancer 2013;109:307-11. [Crossref] [PubMed]
- Takano S, Reichert M, Bakir B, et al. Prrx1 isoform switching regulates pancreatic cancer invasion and metastatic colonization. Genes Dev 2016;30:233-47. [Crossref] [PubMed]
- Chen W, Wu J, Shi W, et al. PRRX1 deficiency induces mesenchymal-epithelial transition through PITX2/miR-200-dependent SLUG/CTNNB1 regulation in hepatocellular carcinoma. Cancer Sci 2021;112:2158-72. [Crossref] [PubMed]
- Du W, Liu X, Yang M, et al. The Regulatory Role of PRRX1 in Cancer Epithelial-Mesenchymal Transition. Onco Targets Ther 2021;14:4223-9. [Crossref] [PubMed]
- Jiang YP, Tang YL, Wang SS, et al. PRRX1-induced epithelial-to-mesenchymal transition in salivary adenoid cystic carcinoma activates the metabolic reprogramming of free fatty acids to promote invasion and metastasis. Cell Prolif 2020;53:e12705. [Crossref] [PubMed]
- Yin L, Liu MX, Wang FY, et al. Transcription Factor prrx1 Promotes Brown Adipose-Derived Stem Cells Differentiation to Sinus Node-Like Cells. DNA Cell Biol 2019;38:1313-22. [Crossref] [PubMed]
- Chen Z, Chen Y, Li Y, et al. Prrx1 promotes stemness and angiogenesis via activating TGF-β/smad pathway and upregulating proangiogenic factors in glioma. Cell Death Dis 2021;12:615. [Crossref] [PubMed]
- Jiang J, Zheng M, Zhang M, et al. PRRX1 Regulates Cellular Phenotype Plasticity and Dormancy of Head and Neck Squamous Cell Carcinoma Through miR-642b-3p. Neoplasia 2019;21:216-29. [Crossref] [PubMed]
- Rebucci M, Michiels C. Molecular aspects of cancer cell resistance to chemotherapy. Biochem Pharmacol 2013;85:1219-26. [Crossref] [PubMed]
- Schlack K, Boegemann M, Steinestel J, et al. The safety and efficacy of gemcitabine for the treatment of bladder cancer. Expert Rev Anticancer Ther 2016;16:255-71. [Crossref] [PubMed]
- de Sousa Cavalcante L, Monteiro G. Gemcitabine: metabolism and molecular mechanisms of action, sensitivity and chemoresistance in pancreatic cancer. Eur J Pharmacol 2014;741:8-16. [Crossref] [PubMed]
- Mini E, Nobili S, Caciagli B, et al. Cellular pharmacology of gemcitabine. Ann Oncol 2006;17:v7-12. [Crossref] [PubMed]
- Barlési F, Villani P, Doddoli C, et al. Gemcitabine-induced severe pulmonary toxicity. Fundam Clin Pharmacol 2004;18:85-91. [Crossref] [PubMed]
- Ichihara N, Kubota Y, Kitanaka A, et al. Inhibition of Src reduces gemcitabine-induced cytotoxicity in human pancreatic cancer cell lines. Cancer Lett 2008;260:155-62. [Crossref] [PubMed]
- Pinto-Leite R, Arantes-Rodrigues R, Palmeira C, et al. Everolimus enhances gemcitabine-induced cytotoxicity in bladder-cancer cell lines. J Toxicol Environ Health A 2012;75:788-99. [Crossref] [PubMed]
- Jones RJ, Baladandayuthapani V, Neelapu S, et al. HDM-2 inhibition suppresses expression of ribonucleotide reductase subunit M2, and synergistically enhances gemcitabine-induced cytotoxicity in mantle cell lymphoma. Blood 2011;118:4140-9. [Crossref] [PubMed]
- Yang G, Li Z, Dong L, et al. lncRNA ADAMTS9-AS1 promotes bladder cancer cell invasion, migration, and inhibits apoptosis and autophagy through PI3K/AKT/mTOR signaling pathway. Int J Biochem Cell Biol 2021;140:106069. [Crossref] [PubMed]
- Zhou X, Chen Y, Wang F, et al. Artesunate induces autophagy dependent apoptosis through upregulating ROS and activating AMPK-mTOR-ULK1 axis in human bladder cancer cells. Chem Biol Interact 2020;331:109273. [Crossref] [PubMed]
- Cheng L, Wang H, Wang Z, et al. Leflunomide Inhibits Proliferation and Induces Apoptosis via Suppressing Autophagy and PI3K/Akt Signaling Pathway in Human Bladder Cancer Cells. Drug Des Devel Ther 2020;14:1897-908. [Crossref] [PubMed]
- Hirata H, Sugimachi K, Takahashi Y, et al. Downregulation of PRRX1 Confers Cancer Stem Cell-Like Properties and Predicts Poor Prognosis in Hepatocellular Carcinoma. Ann Surg Oncol 2015;22:S1402-9. [Crossref] [PubMed]
- An Q, Zhou L, Xu N. Long noncoding RNA FOXD2-AS1 accelerates the gemcitabine-resistance of bladder cancer by sponging miR-143. Biomed Pharmacother 2018;103:415-20. [Crossref] [PubMed]
- Xie F, Zhao N, Zhang H, et al. Circular RNA CircHIPK3 Promotes Gemcitabine Sensitivity in Bladder Cancer. J Cancer 2020;11:1907-12. [Crossref] [PubMed]
- Marchand B, Pitarresi JR, Reichert M, et al. PRRX1 isoforms cooperate with FOXM1 to regulate the DNA damage response in pancreatic cancer cells. Oncogene 2019;38:4325-39. [Crossref] [PubMed]
- Hamurcu Z, Delibaşı N, Nalbantoglu U, et al. FOXM1 plays a role in autophagy by transcriptionally regulating Beclin-1 and LC3 genes in human triple-negative breast cancer cells. J Mol Med (Berl) 2019;97:491-508. [Crossref] [PubMed]
- Chandrasekar T, Evans CP. Autophagy and urothelial carcinoma of the bladder: A review. Investig Clin Urol 2016;57:S89-97. [Crossref] [PubMed]
- Klionsky DJ. Autophagy revisited: a conversation with Christian de Duve. Autophagy 2008;4:740-3. [Crossref] [PubMed]
- Mowers EE, Sharifi MN, Macleod KF. Functions of autophagy in the tumor microenvironment and cancer metastasis. FEBS J 2018;285:1751-66. [Crossref] [PubMed]
- Han Y, Fan S, Qin T, et al. Role of autophagy in breast cancer and breast cancer stem cells Int J Oncol 2018;52:1057-70. (Review). [Crossref] [PubMed]
- Wang F, Tang J, Li P, et al. Chloroquine Enhances the Radiosensitivity of Bladder Cancer Cells by Inhibiting Autophagy and Activating Apoptosis. Cell Physiol Biochem 2018;45:54-66. [Crossref] [PubMed]
- Kou B, Liu W, Xu X, et al. Autophagy induction enhances tetrandrine-induced apoptosis via the AMPK/mTOR pathway in human bladder cancer cells. Oncol Rep 2017;38:3137-43. [Crossref] [PubMed]
- Hung SY, Chen WF, Lee YC, et al. Rhopaloic acid A induces apoptosis, autophagy and MAPK activation through ROS-mediated signaling in bladder cancer. Phytomedicine 2021;92:153720. [Crossref] [PubMed]
- Fu Q, Gao Y, Yang F, et al. Suppression of microRNA-454 impedes the proliferation and invasion of prostate cancer cells by promoting N-myc downstream-regulated gene 2 and inhibiting WNT/β-catenin signaling. Biomed Pharmacother 2018;97:120-7. [Crossref] [PubMed]
- Guilbert SM, Lambert H, Rodrigue MA, et al. HSPB8 and BAG3 cooperate to promote spatial sequestration of ubiquitinated proteins and coordinate the cellular adaptive response to proteasome insufficiency. FASEB J 2018;32:3518-35. [Crossref] [PubMed]
- Sun T, Li Y, Li T, et al. JIP1 and JIP3 cooperate to mediate TrkB anterograde axonal transport by activating kinesin-1. Cell Mol Life Sci 2017;74:4027-44. [Crossref] [PubMed]
- Marivin A, Garcia-Marcos M. DAPLE and MPDZ bind to each other and cooperate to promote apical cell constriction. Mol Biol Cell 2019;30:1900-10. [Crossref] [PubMed]
- Block CJ, Mitchell AV, Wu L, et al. RNA binding protein RBMS3 is a common EMT effector that modulates triple-negative breast cancer progression via stabilizing PRRX1 mRNA. Oncogene 2021;40:6430-42. [Crossref] [PubMed]


