Differential expression of DNA damage repair genes after chemoradiotherapy and inhibition rate in different bladder cancer cells
Introduction
Bladder cancer is the ninth most common type of cancer, often diagnosed at a late stage. Few novel prevention and treatment strategies have been developed for bladder cancer (1), and survival rates have not improved substantially in the past three decades. The bladder is a vital organ, and bladder removal surgery negatively affects quality of life, especially for elderly and frail patients. Conservative treatment can be a good option, but it must contend with the problem of a high recurrence rate. Chemotherapy combined with radiotherapy has been shown to significantly improve locoregional control of bladder cancer, with no significant increase in side effects compared to radiotherapy alone (2). The 2-year survival rate for bladder-sparing therapies with radiation alone or chemoradiotherapy is 42% vs. 56%, respectively (3). However, the latest treatment guidelines state that radiotherapy or chemotherapy alone should not be used as the primary treatment for bladder cancer because residual tumor cells are the leading cause of tumor recurrence (4). Trimodality treatment combines transurethral resection of the bladder tumor, chemotherapy, and radiation, effectively inhibiting cancer recurrence (5,6). Nevertheless, some patients do not respond well to chemoradiotherapy. Few studies focus the cause and mechanism of the chemoradiotherapy resistance in the bladder cancer, related to apoptosis, cell proliferation, DNA damage repair genes, molecular subtype, and others. Determining a patient’s sensitivity to treatment is difficult due to the complexity and heterogeneity of tumors; thus, further research is necessary to understand the mechanisms underlying chemoradiotherapy and to identify the genes involved. DNA damage repair genes are closely associated with the efficacy of radiotherapy and chemotherapy (7,8). These genes help us identify patients most likely to respond to chemoradiotherapy. Furthermore, research along these lines may indicate whether patients who are insensitive to chemoradiotherapy may benefit from combined treatment in the future. Herein, we found some different DNA damage repair genes respond in chemoradiotherapy resistance in the bladder cancer cell lines and identified the olaparib, poly (ADP-ribose) polymerase (PARP) inhibitor, can leading to the tumor cell death by blocking mutagenic on-homologous end joining (NHEJ) repair pathway in high chemoradiotherapy response cell line. We present the following article in accordance with the MDAR reporting checklist (available at https://tau.amegroups.com/article/view/10.21037/tau-22-543/rc).
Methods
Instruments and materials
Irradiation radiotherapy apparatus (Rad Source Technologies, Germany), a Hieff UNICON Universal Blue qPCR SYBR Green Master Mix (Yeassen, Shanghai, China), a Cell Counting Kit-8 (MedChemExpress, Wuhan, China), TRIzol reagent (TaKaRa, Shiga, Japan), a PrimeScript® RT reagent kit (Thermo Fisher, Suzhou, China), olaparib (O126162, Aladdin, China), and a centrifuge (Thermo Fisher Scientific, China) were used for the study. Gemcitabine was a generous gift from Professor Lanlan Tang, Department of Urology, Guizhou Provincial People’s Hospital, Guiyang, China. The cell lines were purchased from American Type Culture Collection (ATCC).
Assessment of the sensitivity of bladder cancer cells to common clinical chemotherapy drugs and radiotherapy
Three bladder cancer cell lines (5637, TCCSUP, and J82) were selected for the experimental treatment. A negative control group, drug concentration gradient chemotherapy group, radiotherapy group, and drug concentration gradient chemotherapy combined with radiotherapy group were established. After culturing the three cell lines to the logarithmic phase, the cells were suspended in 2.5% trypsin and then counted using a cell counter. The 5637, TCCSUP, and J82 cell lines were cultured into 96-well plates with 10,000, 60,000, and 300,000 cells/well, respectively. After 8 h of incubation, the cells were treated with different concentrations of gemcitabine (5, 1, 0.5, 0.1, and 0.01 µM). After 2 h of treatment, the radiotherapy group and the gemcitabine chemotherapy combined with radiotherapy group were treated with radiation at a dose of 9 Gy. The cells were then returned to the incubator for 48 h after irradiation, followed by the addition of CCK8 reagent and a 2-hour wait.
Analysis of radiosensitive bladder cancer genes using information from The Cancer Genome Atlas (TGCA) and other databases and their correlation with overall survival (OS)
The underlying mechanism and changes in genes related to radiosensitivity were investigated further based on the results of clinical trials using common chemotherapy drugs combined with radiotherapy on three different bladder cancer cell lines (5637, TCCSUP, and J82). Zhu et al. used the CRISPR/Cas9 SAM human lentiviral library, which contains 70,290 unique sgRNA sequences targeting 23,430 human genes, to screen the target genes related to radiotherapy sensitivity in neuroblastoma (9). Lan et al. also used the CRISPR/Cas9 library to screen genes related to radiotherapy sensitivity in pancreatic cancer (10). We referred to the database information provided by these two previous studies, combined with the TGCA database, to analyze and identify the possible radioresistance-related genes in bladder cancer cells and the relationship between their expression and bladder cancer survival and prognostic indicators. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Effect of gemcitabine chemotherapy and radiotherapy on the expression levels of related genes in bladder cancer cells
The expression levels of the EXO1, BLM, RBBP8, and BRCA1 genes were detected using the reverse transcription quantitative PCR (RT-qPCR) technique after three bladder cancer cell lines (5637, TCCSUP, and J82) were treated with gemcitabine (the most popular chemotherapy agent). A negative control group, gemcitabine 50% inhibitory concentration (IC50) chemotherapy group, radiotherapy group, and gemcitabine chemotherapy combined with radiotherapy group were established. Based on the growth rate of the three cell lines, the cells were subcultured in 6-well plates. After confluence, the three cell lines were treated with gemcitabine, and the IC50 values were detected for the three cell lines. After 2 h of treatment, the radiotherapy group and the gemcitabine chemotherapy combined with radiotherapy group were treated with radiation at a dose of 9 Gy. An RNA extraction kit and PCR system reagents were used for RNA extraction, reverse transcription, and PCR detection.
Assessment of sensitivity to olaparib plus gemcitabine chemotherapy combined with radiotherapy in bladder cancer cells
The sensitivity of the three bladder cancer cell lines (5637, TCCSUP, and J82) to gemcitabine IC50 combined with radiotherapy was determined based on the expression of EXO1, BLM, RBBP8, and BRCA1 genes in bladder cancer cells. The sequences of the primers used in this study were showed in the Table 1. We established the experimental groups for each cell line based on the recommended dose of olaparib as follows: Negative control group, 5 nM olaparib alone treatment group, bladder cancer cell line gemcitabine IC50 alone treatment group, 5 nM olaparib plus bladder cancer cell line gemcitabine IC50 treatment group, 10 nM olaparib plus bladder cancer cell line gemcitabine IC50 treatment group, and drugs combined with the radiotherapy group. After culturing the three cell lines to the logarithmic phase, 2.5% trypsin was used for cell suspension, and the cells were counted using a cell counter. Then, the 5637, TCCSUP, and J82 cells were cultured into 96-well plates with 10,000, 60,000, and 300,000 cells/well, respectively. After 8 h, the cells were treated with the various drugs. After 2 h of drug treatment, all the experimental groups with radiation therapy were treated with radiation at a dose of 9 Gy. After irradiation, the cells were put back into the incubator and cultured for 48 h. The CCK8 reagent mixed with the culture medium was added, and after 2 h, the absorbance readings were analyzed with an enzyme labeling instrument.
Table 1
| Gene | Primer (5'–3') |
|---|---|
| BLM | Forward: GCT AGA GGC GAT CAA TGC |
| Reverse: GAC CCC AGG AGA AAC ACA | |
| BRCA1 | Forward: TGT TGA TGT GGA GGA GCA |
| Reverse: TTC CAG GTA AGG GGT TCC | |
| EXO1 | Forward: AAC AAG GTT CCT GGG CTA T |
| Reverse: TGC TTT CTC TTC TGG ATG C | |
| RBBP8 | Forward: TGA TCG GAC AAC ACA TG |
| Reverse: CGC TTT CTG CTT GAC TTT |
Statistical analysis
All experiments were repeated in triplicate in the three biological replicates, and the experimental data are expressed as the mean ± standard deviation. SPSS 22.0 statistical software was used for the data analysis. Comparisons among more than two groups were performed using a one-way analysis of variance, and the t-test was used for comparisons between two groups. A P value <0.05 was used to indicate statistical significance. No statistical method was used to predetermine sample size, and no samples and data points were excluded in any analysis.
Results
Enhancement of the lethality effect of combined radiotherapy and chemotherapy on bladder cancer cells
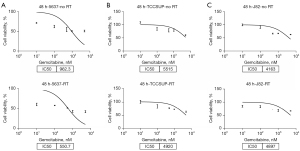
Three bladder cancer cell lines (5637, J82, and TCCSUP) were selected for the experiment. Gemcitabine, a commonly used drug in clinical chemotherapy, was used for the treatment. Each group was subjected to either radiation or no treatment. We found that the combination of gemcitabine and radiotherapy did not significantly increase the inhibition of J82 and TCCSUP cell proliferation but significantly increased the inhibition of 5637 cell proliferation (Figure 1). The 5637 cell line was the most sensitive, and the IC50 values were 962.3 nM for gemcitabine alone and 550.7 nM for gemcitabine combined with radiotherapy. The inhibition increased by 42.8% [(962.3–550.7)/962.3]. The TCCSUP cell line was the second most sensitive, with an IC50 value of 5,515 nM for gemcitabine alone and 4,920 nM for gemcitabine combined with radiotherapy. The inhibition was increased by only 10.8% [(5,515–4,920)/5,515]. The J82 cell line was the least sensitive, with an IC50 value of 4,163 nM for gemcitabine alone and 4,897 nM for gemcitabine combined with radiotherapy. The inhibition decreased by 17.6% [(4,163–4,897)/4,163]. The sensitivity of different bladder cancer cell lines to chemoradiotherapy differed based on the change in gene expression profiles.
Analysis of genes sensitive to chemoradiotherapy and their correlation with OS in bladder cancer
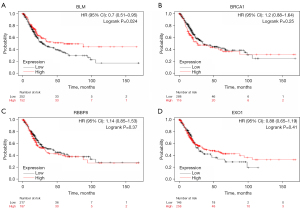
Chemotherapy combined with radiotherapy can improve tumor inhibition, but different cell lines have different sensitivity to chemotherapy. The mechanism behind the sensitivity change was studied by identifying the genes associated with chemoradiotherapy sensitivity. Based on the information in this database, we found that genes related to DNA damage repair were associated with radiation and chemotherapy resistance. The genes related to radiation and chemotherapy resistance in bladder cancer were predicted using the TGCA database. We found four genes (BLM, BRCA1, EXO1, and RBBP8) that significantly correlated with resistance. BLM and EXO1 were involved in DNA mismatch repair (MMR), and BRCA1 and RBBP8 were involved in DNA damage double-strand break repair (DSBR). We found that the expression levels of BLM and EXO1 were correlated with the OS of patients with bladder cancer. When BLM and EXO1 were highly expressed, the OS of patients was prolonged. The expression levels of BRCA1 and RBBP8 were not significantly correlated with the OS of patients with bladder cancer (Figure 2). However, BRCA1 and RBBP8 were correlated with the efficacy of cisplatin-based neoadjuvant chemotherapy, which was more significantly associated with poor disease-free survival (11). BRCA1 mRNA expression was related to the outcome of neoadjuvant cisplatin-based chemotherapy in bladder cancer; the median survival was 168 months for patients with low expression levels and 34 months for patients with high expression levels (12).
Expression levels of related genes after radiotherapy and chemotherapy
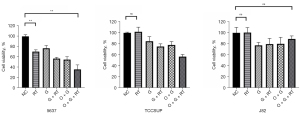
EXO1, BLM, RBBP8, and BRCA1 gene expression levels were detected by qPCR in three different bladder cancer cell lines after different treatments (Figure 3). The three cell lines have different molecular subtypes; 5637 cells are basal, whereas J82 and TCCSUP cells are both non-basal and non-luminal (13). We found that the expression levels of related genes in 5637 cells did not alter significantly after radiotherapy and chemotherapy, but the expression levels of BRCA1 and RBBP8 were significantly increased in J82 and TCCSUP cells. A high expression of BRCA1 and RBBP8 induces resistance to radiation and chemotherapy by activating the homologous recombination (HR) pathway of DNA damage repair.

Inhibitory effect and expression of related genes after treatment combined with olaparib
Olaparib is a PARP inhibitor that plays a crucial role in initiating DNA repair. This drug is approved by the Food and Drug Administration (FDA) for treating BRCA-mutated triple-negative breast cancer. It exploits BRCA mutation deficiency through a synthetic lethality and is considered promising for anticancer therapy. The reactive expression of BRCA1 and RBBP8 in these three cell lines differed after radiotherapy and chemotherapy. Combined treatment with olaparib resulted in a significant increase in the inhibition efficiency against 5637 cells, whereas the inhibitory efficiency against J82 cells was decreased (Figure 4).
Discussion
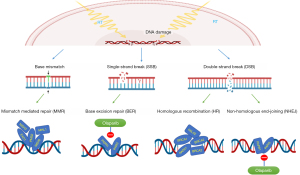
MMR is a repair pathway that restores the normal nucleotide sequence in a DNA molecule and is used to correct mismatched base pairs in the DNA double helix. MMR deficiency is the primary mechanism of cisplatin/carboplatin resistance. MMR mutations can occur in cisplatin-resistant cells (14). A recent study identified the involvement of MMR in drug resistance by RNA interference (RNAi) knockdown and CRISPR/Cas9 knockout screenings. MMR interacts with DNA damage in platinum-based chemotherapy, leading to tumor cell death. On the other hand, loss of MMR-related proteins can be used to identify patients who would benefit from radiotherapy (14). Radiation therapy uses high-energy radiation to damage DNA in tumor cells. MMR is involved in the DNA repair process and affects sensitivity to radiotherapy. Hence, the effect of MMR on the sensitivity to radiotherapy and chemotherapy is the opposite. The BLM gene is involved in the complex formation of PMS2, EXO1, and PMS2, which are MMR-related proteins (Figure 5). We observed that the change in EXO1 and BLM expression levels was not as significant as that in BRCA1 and RBBP8 expression levels after treatment with combined radiotherapy and chemotherapy.
In addition to MMR, other DNA break repair mechanisms, including single-strand break repair and DSBR, are also involved. DSBR is divided into NHEJ and HR. RBBP8, also known as C terminal binding protein-interacting protein, is used not only for repair of DSBR by HR in the S/G2 phase but also for microhomology-mediated end joining in G1 (15). RBBP8 is highly expressed in tumor cells and affects the prognosis of patients with malignancies, including plasma cell myeloma, gliomas, and nasopharyngeal malignancies (16,17). RBBP8 recruits the breast cancer susceptibility gene 1 (BRCA1) in HR (18). BRCA1 is mainly involved in the HR of DNA damage (Figure 5). BRCA1 plays a core role in HR, and BRCA1 expression is associated with sensitivity to cisplatin-based chemotherapy in bladder cancer (19). A low expression of BRCA1 induces DNA damage by repressing the HR pathway and sensitizes tumor cells to radiation and chemotherapy. We found that the sensitivity of the three cell lines to radiotherapy and chemotherapy was different; the 5637 cell line was the most sensitive, followed by the TCCSUP and J82 cell lines. The expression levels of related genes in 5637 cells did not significantly change after radiotherapy and chemotherapy, while the expression levels of BRCA1 and RBBP8 significantly increased in J82 and TCCSUP cells. This result is also consistent with the fact that BRCA1 and RBBP8 form a complex that initiates the HR repair pathway, leading to decreased sensitivity of tumor cells to treatment.
When analyzing the DNA damage repair pathways, we found two other pathways: base excision repair (BER) and NHEJ (Figure 5). PARP1 plays a crucial role in these two DNA damage repair pathways (20). Olaparib, a PARP inhibitor, is an FDA-approved treatment for BRCA-mutated triple-negative breast cancer (21) and metastatic castration-resistant prostate cancer (22). Olaparib exploits BRCA mutation deficiency through a synthetic lethality and is considered a promising anticancer therapy. If tumor cells harbor BRCA mutations, they must activate the BER or NHEJ pathways to repair DNA. Using PARP inhibitors can completely inhibit the damage repair function of tumor cells, resulting in synthetic lethality (23). However, normal cells without BRCA mutations can repair damaged DNA through the HR pathway. In our experiment, the expression levels of BRCA1 and RBBP8 did not increase significantly after treating 5637 cells with radiotherapy and chemotherapy, suggesting insensitive activation of the HR pathway. The inhibition efficiency against 5637 cells was improved considerably after adding olaparib to the therapy, and the opposite result was observed in J82 cells. The expression levels of BRCA1 and RBBP8 in J82 cells were significantly increased after chemoradiotherapy. After adding olaparib to the therapy, J82 cells reduced the inhibitory efficiency of chemoradiotherapy. Olaparib was ineffective due to its inability to induce synthetic lethality, as suggested by the high reactive expression of BRCA1 and RBBP8.
In conclusion, the expression levels of RBBP8 and BRCA1 were correlated with sensitivity to radiotherapy and chemotherapy in bladder cancer; the higher the reactive expression after treatment, the worse the sensitivity. As such, the reactive expression levels of BRCA1 and RBBP8 after chemoradiotherapy may be useful in evaluating the efficacy of olaparib combination therapy.
Acknowledgments
Funding: This work was supported by the Natural Science Foundation of Jiangxi Province of China (No. 20202BABL216060).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://tau.amegroups.com/article/view/10.21037/tau-22-543/rc
Data Sharing Statement: Available at https://tau.amegroups.com/article/view/10.21037/tau-22-543/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tau.amegroups.com/article/view/10.21037/tau-22-543/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Grayson M. Bladder cancer. Nature 2017;551:S33. [Crossref] [PubMed]
- James ND, Hussain SA, Hall E, et al. Radiotherapy with or without chemotherapy in muscle-invasive bladder cancer. N Engl J Med 2012;366:1477-88. [Crossref] [PubMed]
- Korpics MC, Block AM, Martin B, et al. Concurrent chemotherapy is associated with improved survival in elderly patients with bladder cancer undergoing radiotherapy. Cancer 2017;123:3524-31. [Crossref] [PubMed]
- Witjes JA, Bruins HM, Cathomas R, et al. European Association of Urology Guidelines on Muscle-invasive and Metastatic Bladder Cancer: Summary of the 2020 Guidelines. Eur Urol 2021;79:82-104. [Crossref] [PubMed]
- Yoshioka H, Shimbo T, Yoshida K, et al. Treatment Results of Radiotherapy Combined with Balloon-occluded Arterial Infusion Chemotherapy for Invasive Bladder Cancer. Anticancer Res 2016;36:731-6. [PubMed]
- Patel A, Spychalski P, Kobiela J. Short-course radiotherapy and consolidation chemotherapy: in search of a balanced protocol. Br J Surg 2022;109:e50-1. [Crossref] [PubMed]
- Lozinski M, Bowden NA, Graves MC, et al. Transcriptomic Profiling of DNA Damage Response in Patient-Derived Glioblastoma Cells before and after Radiation and Temozolomide Treatment. Cells 2022;11:1215. [Crossref] [PubMed]
- Ouellette MM, Zhou S, Yan Y. Cell Signaling Pathways That Promote Radioresistance of Cancer Cells. Diagnostics (Basel) 2022;12:656. [Crossref] [PubMed]
- Zhu GD, Yu J, Sun ZY, et al. Genome-wide CRISPR/Cas9 screening identifies CARHSP1 responsible for radiation resistance in glioblastoma. Cell Death Dis 2021;12:724. [Crossref] [PubMed]
- Lan B, Zeng S, Zhang S, et al. CRISPR-Cas9 Screen Identifies DYRK1A as a Target for Radiotherapy Sensitization in Pancreatic Cancer. Cancers (Basel) 2022;14:326. [Crossref] [PubMed]
- Herrmann J, Schmidt H, Nitschke K, et al. RNA Expression of DNA Damage Response Genes in Muscle-Invasive Bladder Cancer: Influence on Outcome and Response to Adjuvant Cisplatin-Based Chemotherapy. Int J Mol Sci 2021;22:4188. [Crossref] [PubMed]
- Font A, Taron M, Gago JL, et al. BRCA1 mRNA expression and outcome to neoadjuvant cisplatin-based chemotherapy in bladder cancer. Ann Oncol 2011;22:139-44. [Crossref] [PubMed]
- Silina L, Maksut F, Bernard-Pierrot I, et al. Review of Experimental Studies to Improve Radiotherapy Response in Bladder Cancer: Comments and Perspectives. Cancers (Basel) 2020;13:87. [Crossref] [PubMed]
- Reijnen C, Küsters-Vandevelde HVN, Prinsen CF, et al. Mismatch repair deficiency as a predictive marker for response to adjuvant radiotherapy in endometrial cancer. Gynecol Oncol 2019;154:124-30. [Crossref] [PubMed]
- Davies OR, Forment JV, Sun M, et al. CtIP tetramer assembly is required for DNA-end resection and repair. Nat Struct Mol Biol 2015;22:150-7. [Crossref] [PubMed]
- Liu Z, Cheng X, Lin S, et al. Prognostic Significance of mRNA Expression RBBP8 or Its Methylation in Gliomas. Cell Mol Neurobiol 2022; Epub ahead of print. [Crossref] [PubMed]
- Zhang W, Song Y, He X, et al. Prognosis value of RBBP8 expression in plasma cell myeloma. Cancer Gene Ther 2020;27:22-9. [Crossref] [PubMed]
- Yun MH, Hiom K. CtIP-BRCA1 modulates the choice of DNA double-strand-break repair pathway throughout the cell cycle. Nature 2009;459:460-3. [Crossref] [PubMed]
- Fu X, Tan W, Song Q, et al. BRCA1 and Breast Cancer: Molecular Mechanisms and Therapeutic Strategies. Front Cell Dev Biol 2022;10:813457. [Crossref] [PubMed]
- Hu X, Zhang J, Zhang Y, et al. Dual-target inhibitors of poly (ADP-ribose) polymerase-1 for cancer therapy: Advances, challenges, and opportunities. Eur J Med Chem 2022;230:114094. [Crossref] [PubMed]
- Robert M, Frenel JS, Gourmelon C, et al. Olaparib for the treatment of breast cancer. Expert Opin Investig Drugs 2017;26:751-9. [Crossref] [PubMed]
- Dong X, Liu HJ, Feng HY, et al. Enhanced Drug Delivery by Nanoscale Integration of a Nitric Oxide Donor To Induce Tumor Collagen Depletion. Nano Lett 2019;19:997-1008. [Crossref] [PubMed]
- Waissi W, Nicol A, Jung M, et al. Radiosensitizing Pancreatic Cancer with PARP Inhibitor and Gemcitabine: An In Vivo and a Whole-Transcriptome Analysis after Proton or Photon Irradiation. Cancers (Basel) 2021;13:527. [Crossref] [PubMed]
(English Language Editor: D. Fitzgerald)