Prevalence of germline mutations in cancer susceptibility genes in Chinese patients with renal cell carcinoma
Highlight box
Key findings
• Pathogenic germline mutations in FANCI and FANCM in RCC; heterozygous germline missense mutation in exon 5 of the FH gene c.563A>T:p.N188I in RCC.
What is known and what is new?
• Previous research found that germline mutations in VHL, FH, SDHB, SDHC, SDHD, MET, FLCN, PTEN, TSC1, TSC2, MITF, BAP1, PBRM1, and CDKN2B can increase the risk of RCC.
• We found that germline mutations of the FANCI gene c.158-2A>G and FANCM gene c.4515+1G>C exist in patients with RCC; heterozygous germline missense mutation in exon 5 of the FH gene c.563A>T:p.N188I in RCC.
What is the implication, and what should change now?
• FANCI and FANCM pathogenic germline mutations might play an important role in tumorigenesis and tumor progression in RCC. Young RCC patients, patients with bilateral or multifocal RCC, or patients with nccRCC should be offered genetic testing for more precise clinical implications.
Introduction
Renal cell carcinoma (RCC) is the ninth most frequently diagnosed cancer in men (1). RCC comprises several subtypes, including clear cell RCC (ccRCC), papillary RCC type 1 (pRCC T1), pRCC T2, mixed oncocytoma or chromophobe RCC (chRCC), and other rare subtypes (2). Tumors that do not meet the criteria for any established subtype are categorized as unclassified (3). Loss of function of the von Hippel-Lindau (VHL) protein, mutations in MET, FH, and FLCN have been considered major mutations in ccRCC, pRCC T1, and pRCC T2, and mixed oncocytoma or chRCC, respectively (2). Germline mutations, one of genetic alterations, exist in all cells in body and can be inherited (4,5). Germline mutations are associated with generation of various tumors and drug resistance (6-8). A recent American study found that 16.1% of RCC patients carried germline mutations in a RCC predisposition gene, and some cases of early-onset aggressive RCC without defined pathogenic germline mutations have been observed (9). It seems highly probable that underlying causative germline mutations exist in RCC. Thus, further investigations need to be explored in different patient populations with special molecular and clinical characteristics.
A previous report suggested that patients with renal cancer affected by hereditary factors account for 3–5% of all RCC patients (10), which is likely underestimated (11). Studies have shown that germline mutations in 14 genes (VHL, FH, SDHB, SDHC, SDHD, MET, FLCN, PTEN, TSC1, TSC2, MITF, BAP1, PBRM1, and CDKN2B) can increase the risk of RCC (12-15). It is particularly relevant to identify Chinese patients with inherited RCC because their clinical implications can differ from those of patients with sporadic RCC (16,17). However, the genetic basis of some inherited renal cancers has not been clearly elucidated.
To explore the genetic basis of inherited renal cancers, we conducted a comprehensive germline analysis of multiple renal cancer predisposition genes in a cohort of Chinese patients. Among the patients in our cohort, 10.57% carried pathogenic/likely pathogenic germline mutations. Most (6/10) of the mutated genes were associated with maintenance of genomic stability and DNA repair. Young RCC patients, patients with bilateral or multifocal RCC, or patients with non-clear cell RCC (nccRCC) are more likely to have pathogenic/likely pathogenic germline mutations. Importantly, this is the first study to report that identical germline mutations of the FANCI gene c.158-2A>G and FANCM gene c.4515+1G>C exist in patients with RCC and is the first article to show heterozygous germline missense mutation in exon 5 of the FH gene c.563A>T:p.N188I in RCC. We present the following article in accordance with the MDAR reporting checklist (available at https://tau.amegroups.com/article/view/10.21037/tau-23-32/rc).
Methods
Patients
Peripheral blood was obtained from 123 patients with RCC in the Department of Urology, The Third Medical Center, Chinese PLA General Hospital between 1 August 2017 and 31 December 2020. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of the Chinese PLA General Hospital (No. S2013-065-01) and informed consent was taken from all the patients.
Genomic sequencing
In our cohort, whole-exome sequencing (WES) (n=69) or gene panel sequencing containing 139 genes (n=54) related to germline cancer predisposition was performed to evaluate the pathogenic/likely pathogenic germline mutation rate and identify de novo pathogenic/likely pathogenic germline mutations in patients with RCC (Table S1). Germline DNA was obtained from peripheral white blood. Polymerase chain reaction (PCR) was performed to amplify DNA. The enriched DNA, which was converted to sequencing libraries, was sequenced and analyzed using the Illumina Novoseq platform (Illumina, San Diego, CA, USA), as previously described (18).
Statistics
Clinical and pathologic characteristics were compared using chi-square test (χ2) and P values <0.05 were considered significant. Statistical analyses were performed using SPSS 24.0 software (IBM Corp., Armonk, NY, USA).
Results
Patient characteristics
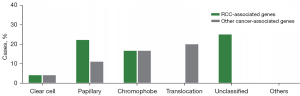
The characteristics and clinical features of the 123 patients are summarized in Table 1. In our study, the age at diagnosis varied from 19 to 85 years (median, 54 years). Among the 123 patients, 95 had ccRCC (77.2%) and 28 had nccRCC (22.8%). Overall, 7 patients (5.7%) with a history of a second malignant tumor were identified. Of the 7 patients with secondary malignant tumors, 2 were diagnosed with breast cancer (data not shown). A total of 16 (13.0%) patients with bilateral or multifocal disease were reported and only 2 (1.6%) patients had a family history of RCC (data not shown).
Table 1
| Characteristics | N | % |
|---|---|---|
| Total | 123 | 100.0 |
| Age at diagnosis (years) | ||
| Median | 54 | |
| Range | 19 to 85 | |
| Age 46 or younger | 41 | 33.3 |
| Age 47 or older | 82 | 66.7 |
| Sex | ||
| Female | 30 | 24.4 |
| Male | 93 | 75.6 |
| Race or ethnic background | ||
| Chinese | 123 | 100.0 |
| Family history of RCC | ||
| First-degree relative | 1 | 0.8 |
| First, second, or third-degree relative | 2 | 1.6 |
| More than 1 relative | 2 | 1.6 |
| Family history of malignancy | ||
| First-degree relative | 34 | 27.6 |
| First, second or third-degree relative | 39 | 31.7 |
| More than 1 relative | 11 | 8.9 |
| Tumor histologic subtype | ||
| Clear cell | 95 | 77.2 |
| Papillary | 9 | 7.3 |
| Chromophobe | 6 | 4.9 |
| Translocation-associated | 6 | 4.9 |
| Unclassified | 4 | 3.3 |
| Others | 3 | 2.4 |
| Patient history of prior malignancy | ||
| Yes | 7 | 5.7 |
| No | 116 | 94.3 |
| Bilateral or multifocal RCC at diagnosis | ||
| Yes | 16 | 13.0 |
| No | 107 | 87.0 |
| More than one germline mutation | ||
| Yes | 2 | 1.6 |
| No | 121 | 98.4 |
| Nephrectomy | ||
| Yes | 121 | 98.4 |
| No | 2 | 1.6 |
| Stage at diagnosis | ||
| Stage I | 52 | 42.3 |
| Stage II | 5 | 4.1 |
| Stage III | 41 | 33.3 |
| Stage IV | 23 | 18.7 |
| Unknown | 2 | 1.6 |
| Grade at diagnosis | ||
| Grade I | 7 | 5.7 |
| Grade II | 52 | 42.3 |
| Grade III | 34 | 27.6 |
| Grade IV | 5 | 4.1 |
| Unknown | 25 | 20.3 |
RCC, renal cell carcinoma.
Germline mutations
A total of 13 patients (10.6%) harbored pathogenic or likely pathogenic germline variants in 10 different cancer predisposition genes (Figure 1A). Mutations in RCC-associated genes were identified in 9 (7.3%) patients, and mutations in genes not clearly associated with RCC were identified in 6 (4.9%) patients (Figure 1B). A total of 6 of 95 (6.3%) ccRCC patients had pathogenic/likely pathogenic germline mutations, and 7 of 28 (25.0%) nccRCC patients had pathogenic/likely pathogenic germline mutations (Figure 1C, Table 2). Among the 95 ccRCC patients, 4 (4.2%) patients harbored pathogenic/likely pathogenic germline mutations in RCC-associated genes (Figure 1D). Among the 28 patients with nccRCC, 5 (17.9%) harbored pathogenic/likely pathogenic germline mutations in RCC-associated genes (Figure 1D). For RCC-associated genes (Figure 1B, Table S2), 4 deleterious VHL mutations were detected in 4 individuals (3.3%), including 75.0% (n=3) known deleterious missense mutations and 25.0% (n=1) nonsense mutations. We detected 2 deleterious FLCN mutations in 2 participants (1.6%), including 1 frameshift in/del mutation and 1 deleterious missense mutation. We detected 2 deleterious FH mutations in 2 participants (1.6%), both of which were deleterious missense mutations. We also found 1 SDHB deleterious missense mutation in 1 participant (0.8%). For other cancer-associated genes (Figure 1B, Table S2), 1 MUTYH deleterious missense mutation, 1 RAD51C deleterious missense mutation, 1 NBN likely pathogenic missense mutation, 1 RAD50 frameshift mutation, 1 FANCI deleterious missense mutation, and 1 FANCM deleterious missense mutation were identified. The VHL, FH, FLCN, SDHB, MUTYH, NBN, and RAD51C pathogenic or likely pathogenic mutations in RCC have been reported before, whereas the FANCI and FANCM pathogenic germline mutations in RCC were reported for the first time in this study. Interestingly, 2 patients in our cohort had simultaneous occurrences of 2 causative mutations (Table S2). Patients harboring VHL and FANCM mutations (nonsense and missense, respectively) simultaneously experienced stage I ccRCC at 29 years of age, without a family history of malignant diseases. Similar characteristics were observed in a patient harboring VHL and RAD51C mutations (deletions and missense mutations, respectively). He was diagnosed with stage I ccRCC at 44 years of age, with a family history of malignant cancers. Compared with the phenotypes of patients with only 1 mutation in VHL, no apparent phenotypic differences were found between these 2 patients. The only difference was that patients with only VHL mutations had a single lesion, whereas the patients with both VHL and FANCM/RAD51C mutations had multifocal RCC. In these 2 cases, it seemed that additional FANCM or RAD51C mutations did not affect the age of RCC onset. Thus, to draw a decisive conclusion, further investigations are required to demonstrate the precise role of FANCM and RAD51C in tumorigenesis and tumor progression of RCC.
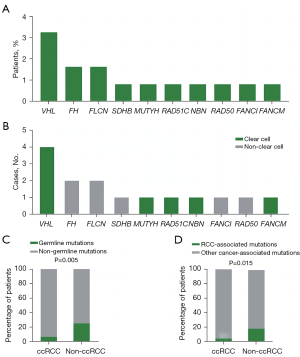
Table 2
| Clinicopathologic parameters | With germline mutations | Without germline mutations | P value |
|---|---|---|---|
| Age at diagnosis, n | <0.001 | ||
| ≤46 years | 11 | 30 | |
| >46 years | 2 | 80 | |
| Subtype, n | 0.005 | ||
| Clear cell | 6 | 89 | |
| Non-clear cell | 7 | 21 | |
| Stage, n | 0.396 | ||
| Stage I & stage II | 8 | 53 | |
| Stage III & stage IV | 5 | 55 | |
| Grade | 0.678 | ||
| Grade 1 & grade 2 | 6 | 53 | |
| Grade 3 & grade 4 | 3 | 36 | |
| Family history of malignancy, n | 0.369 | ||
| Yes | 6 | 39 | |
| No | 7 | 77 | |
| Bilateral or multifocal RCC, n | 0.004 | ||
| Yes | 5 | 11 | |
| No | 8 | 99 |
RCC, renal cell carcinoma.
Clinical characteristics associated with germline mutations
Our cohort comprised 6 patients with ccRCC, 2 patients with pRCC, 2 with chRCC, 1 with microphthalmia (MiT) family translocation RCC, and 2 with unclassified RCC who carried pathogenic/likely pathogenic germline mutations (Figure 2, Table S2). The proposed renal cancer predisposition gene FH pathogenic germline mutations were identified in the 2 patients with pRCC. There were 2 patients with chRCC who harbored mutations in either FLCN or SDHB. The mean age of onset of RCC in germline mutation carriers was 37.2 years. Of these 13 patients, 2 (15.4%) experienced metastasis. A positive family history of cancer was reported in 6 patients (Table S2). The prevalence of deleterious germline mutations is associated with the age at diagnosis. The probability of germline mutations occurring in patients aged 46 years or younger remained higher than that in patients older than 46 years [11 (26.8%) of 41 vs. 2 (2.4%) of 82, P<0.001] (Table 2). We also found that the prevalence of germline mutations was associated with the histologic subtype. Patients with nccRCC were more likely to carry germline mutations [7 (25%) of 28 vs. 6 (6.3%) of 95, P=0.005] (Figure 1C, Table 2). Furthermore, we also found that patients with nccRCC were more likely to have RCC-associated gene mutations [5 (17.9%) of 28 vs. 4 (4.2%) of 95, P=0.015] (Figure 1D). Compared to patients without bilateral or multifocal RCC, those with bilateral or multifocal RCC were more likely to harbor deleterious mutations [5 (38.5%) of 13 vs. 11 (10.0%) of 110, P=0.004] (Table 2). However, a family history of cancer was not associated with more mutations (Table 2). Additionally, a higher stage or grade was not associated with more deleterious pathogenic germline mutations in our study.
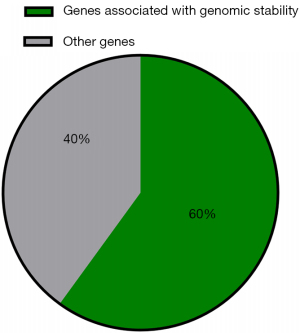
Genes involved in the maintenance of genomic stability or DNA repair were frequently mutated
In our cohort, 13 (13/123, 10.6%) patients harbored deleterious germline mutations in cancer predisposition genes. Some 6 of 10 cancer predisposition genes were associated with maintenance of genomic stability and DNA repair (Figure 3). Deleterious germline mutations of 3 Fanconi anemia (FA)-related genes (FANCI, FANCM, and RAD51C) were detected. The protein products of these genes function cooperatively with other proteins associated with DNA repair processes to maintain genome homeostasis, and loss of function of these genes leads to FA (19). In addition, deleterious germline mutations in RAD50 and NBN have been detected: RAD50 and NBN, members of the MRE11/RAD50/NBN double-strand break repair complex, are thought to repair DNA double-strand breaks and activate DNA damage-induced checkpoints (20,21). Loss of function of these genes is associated with rare chromosomal instability disorders (20,21). Additionally, a deleterious germline mutation in MUTYH was detected. MUTYH, a gene encoding a DNA glycosylase, plays an important role in oxidative DNA damage repair. Inactivation of MUTYH leads to G:C>T:A transversion, which increases the risk of colorectal cancer (22).
Novel pathogenic germline mutation in RCC
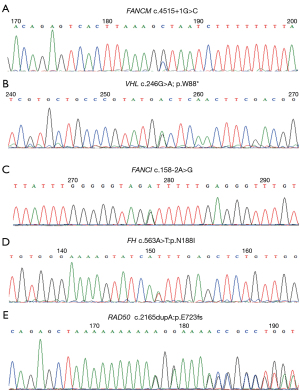
In this study, FANCI and FANCM pathogenic/likely pathogenic germline mutations in patients with RCC were reported for the first time. Patient RCC88 diagnosed with ccRCC carried a FANCM germline mutation annotated as pathogenic in ClinVar (23). This revealed a heterozygous NM_020937:c.4515+1G>C germline FANCM missense mutation, resulting in inactivation of FANCM (Table S3). This intronic variant is a nucleotide substitution located 1 nucleotide downstream of coding exon 17. Interestingly, patient RCC88 simultaneously harbored a pathogenic VHL germline mutation. Subsequent Sanger sequencing of DNA samples collected from the blood of patient RCC88 confirmed the presence of FANCM and VHL germline mutations (Figure 4A,4B). Interestingly, genetic testing of DNA samples collected from the proband’s parents and brother revealed that none of them were carriers of FANCM or VHL mutations (Figure S1), suggesting that the de novo germline mutation occurs during formation of the sperm or egg from an unaffected parent. In addition, patient RCC88 did not have a family history of cancer. Patient RCC63, diagnosed with unclarified RCC, carried a heterozygous NM_001113378: c.158-2A>G germline FANCI missense mutation, which is predicted to inactivate the FANCI gene (Table S3). This intronic variant is a nucleotide substitution located 2 nucleotides upstream of coding exon 4. Subsequent Sanger sequencing of DNA samples collected from the blood of patient RCC63 confirmed FANCI germline mutations (Figure 4C). Annotated as pathogenic in ClinVar, the FANCI variant was considered pathogenic according to the American College of Medical Genetics and Genomics (ACMG) criteria (24). The patient’s father was diagnosed with gastric cancer. In addition, we report a case of aggressive hereditary leiomyomatosis and RCC (HLRCC) in a 32-year-old woman who presented with a novel heterozygous germline missense mutation in exon 5 of FH (c.563A>T:p.N188I) (Figure 4D, Table S3). The patient’s mother was diagnosed with endometrial carcinoma. Consistent with a recent study, we also found a pathogenic RAD50 germline mutation (c.2165dupA; p.E723fs) in a 24-year-old RCC patient with no family history of cancer (Figure 4E, Table S3) (25).
Discussion
Patients with renal cancer affected by hereditary factors account for 3–5% of all RCC patients. Pathogenic germline mutations in cancer predisposition genes are associated with tumorigenesis and development of RCC (26). VHL disease is an autosomal dominant disorder caused by germline mutations in the VHL gene. Patients with VHL disease are particularly prone to renal tumors when a stochastic secondary inactivation of the other VHL allele occurs (27). Similar to those with VHL disease, patients with TSC1/2, FH and SDHB/C/D germline mutations are more likely to have RCC (28-30). Germline mutation in MET can promote hereditary pRCC initiation and progression (31,32). The identification of RCC patients with certain pathogenic germline mutations has important clinical implications, guiding systemic therapy and clinical trial eligibility (16,17,33). Although most studies have investigated the prevalence of germline mutations among patients with RCC and some cancer-predisposition genes involved in the tumorigenesis and tumor progression of RCC have been identified (9,25,34), the genetic basis of some inherited renal cancers has not been clearly elucidated. Here, we conducted a study using WES or gene panel sequencing of 139 genes to evaluate the pathogenic/likely pathogenic germline mutation rate and identify de novo pathogenic/likely pathogenic germline mutations in patients with RCC.
Patients with nccRCC or multifocal RCC, multifocal disease at diagnosis, are at higher risk for inherited syndromes and are significantly more likely to have pathogenic germline mutations (9). Similarly, we showed that patients with bilateral or multifocal nccRCC are more likely to have pathogenic/likely pathogenic germline mutations. Early-onset cancer is a hallmark of an inherited cancer predisposition (35,36), and studies have shown that patients with mutations in RCC-associated genes, such as VHL, FH, FLCN, and SDHB, are at risk for the development of early-onset RCC (37-39). Loss of function of MUTYH accounts for 3% of early-onset CRC (40,41). Mutations in MRE11-RAD50-NBS1 (MRN) complex components, FANCM, or FANCI are associated with early-onset cancers (42-44). We also found that patients with early onset are more likely to carry pathogenic mutations (Table 2). However, Carlo et al. suggested that age at diagnosis was not related to the possibility of germline mutations in patients with advanced RCC (9). These seemingly contradictory findings may be due to Carlo et al. only including patients with advanced tumor stages in their study.
Genome instability is a hallmark of cancer cells and can be accelerated by defects in cellular responses to DNA damage (45,46). In our study, we found that 6 of 10 cancer predisposition genes were associated with maintenance of genomic stability and DNA repair (Figure 3). Deleterious germline mutations of 3 FA-related genes were reported. FA is considered a genetic disease associated with a predisposition to non-hematological and hematological malignancies (47,48). The proteins encoded by these genes comprise the DNA damage response (DDR) system and play vital roles in various cellular processes. Increasing evidence suggests that the defective function of these proteins can be associated with genomic instability, increasing cancer risk (49). Thus, further studies are required to elucidate the role of each component in tumorigenesis and tumor progression in RCC. RAD50 and NBN are components of the MRN complex and are involved in the repair of DNA double-strand breaks. As one of the first sensors and responders to DNA damage, the MRN complex plays a vital role in DDR (45,50). Mutations in the MRN complex are also associated with an increased risk of cancer, suggesting that the complex functions as a tumor suppressor (51-54). However, some studies have shown that the complete knockout of any component of the murine MRN complex can impair early embryonic development (55-57), suggesting that some functions of the MRN complex must be preserved in human disease-associated alleles.
In our study, we found that 2 patients with RCC harbored FANCI and FANCM pathogenic germline mutations. To our knowledge, this is the first report of germline mutations in these pathogenic genes in RCC. As highly conserved DNA remodeling enzymes (58-60), FANCI and FANCM can activate the FA DNA repair pathway to maintain genomic stability. Patients with FA, characterized by the clinically and genetically heterogeneous syndrome of bone marrow failure, are predisposed with a predisposition to cancers (61-64). Studies have shown that the loss of function of some FA genes (such as BRCA1 and BRCA2) by germline inactivation can result in familial breast cancer predisposition syndromes (65-70). In addition, pathogenic FA germline mutations can occur in colorectal cancer, pancreatic cancer, and leukemia (71-73). As members of the FA complementation group, FANCI and FANCM play important roles in the initiation of various cancers. Wang et al. found FANCM mutation in a patient with four primary cancer including renal cancer (74). Mutations in these two genes are associated with renal ectopia malformations or dysplasia (19,75) Thus, we suspected that FANCI and FANCM pathogenic germline mutations might play an important role in tumorigenesis and tumor progression in RCC and further studies are needed to confirm this hypothesis.
There were some limitations in our study. Larger cohorts will need to be studied to confirm the frequency of FANCI and FANCM pathogenic germline mutations in RCC and basic experiments are required to validate the biological role of FANCI and FANCM pathogenic germline mutations in RCC.
Conclusions
Considering our results and the evidence in the literature, the proportion of patients with RCC in our study who carried pathogenic germline mutations may be underestimated. Genetic testing of all patients with nccRCC, patients with bilateral or multifocal RCC, especially those aged 46 years or younger might help identify individual patients for whom targeted therapies are indicated. Patients with FANCI and FANCM pathogenic germline mutations may play an important role in tumorigenesis and tumor progression in RCC.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://tau.amegroups.com/article/view/10.21037/tau-23-32/rc
Data Sharing Statement: Available at https://tau.amegroups.com/article/view/10.21037/tau-23-32/dss
Peer Review File: Available at https://tau.amegroups.com/article/view/10.21037/tau-23-32/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tau.amegroups.com/article/view/10.21037/tau-23-32/coif). Xu Zhang serves as an unpaid editorial board member of Translational Andrology and Urology from April 2022 to March 2023. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of the Chinese PLA General Hospital (No. S2013-065-01) and informed consent was taken from all the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. [Crossref] [PubMed]
- Nguyen KA, Syed JS, Espenschied CR, et al. Advances in the diagnosis of hereditary kidney cancer: Initial results of a multigene panel test. Cancer 2017;123:4363-71. [Crossref] [PubMed]
- Kovacs G, Akhtar M, Beckwith BJ, et al. The Heidelberg classification of renal cell tumours. J Pathol 1997;183:131-3. [Crossref] [PubMed]
- Tischler J, Crew KD, Chung WK. Cases in Precision Medicine: The Role of Tumor and Germline Genetic Testing in Breast Cancer Management. Ann Intern Med 2019;171:925-30. [Crossref] [PubMed]
- Anwar SL, Lehmann U. DNA methylation, microRNAs, and their crosstalk as potential biomarkers in hepatocellular carcinoma. World J Gastroenterol 2014;20:7894-913. [Crossref] [PubMed]
- Park SJ, Armstrong S, Kim CH, et al. Lack of EGF receptor contributes to drug sensitivity of human germline cells. Br J Cancer 2005;92:334-41. [Crossref] [PubMed]
- Li N, Liu C, Xiong L, et al. Pedigree analysis of the EGFR p.V1010M germline mutation in a family with a family history of non-small-cell lung cancer. Ann Transl Med 2022;10:154. [Crossref] [PubMed]
- Urrutia E, Chen H, Zhou Z, et al. Integrative pipeline for profiling DNA copy number and inferring tumor phylogeny. Bioinformatics 2018;34:2126-8. [Crossref] [PubMed]
- Carlo MI, Mukherjee S, Mandelker D, et al. Prevalence of Germline Mutations in Cancer Susceptibility Genes in Patients With Advanced Renal Cell Carcinoma. JAMA Oncol 2018;4:1228-35. [Crossref] [PubMed]
- Lipworth L, Tarone RE, McLaughlin JK. The epidemiology of renal cell carcinoma. J Urol 2006;176:2353-8. [Crossref] [PubMed]
- Yngvadottir B, Andreou A, Bassaganyas L, et al. Frequency of pathogenic germline variants in cancer susceptibility genes in 1336 renal cell carcinoma cases. Hum Mol Genet 2022;31:3001-11. [Crossref] [PubMed]
- Ricketts CJ, Crooks DR, Sourbier C, et al. SnapShot: Renal Cell Carcinoma. Cancer Cell 2016;29:610.610.e1. [Crossref] [PubMed]
- Li J, Liu F, Liu X, et al. Heterozygous germline FLCN mutation in Birt-Hogg-Dubé syndrome with bilateral renal hybrid oncocytic/chromophobe tumor and unilateral renal chromophobe cell carcinoma: a case report. J Cancer Res Clin Oncol 2022; Epub ahead of print. [Crossref] [PubMed]
- Sebai M, Tulasne D, Caputo SM, et al. Novel germline MET pathogenic variants in French patients with papillary renal cell carcinomas type I. Hum Mutat 2022;43:316-27. [Crossref] [PubMed]
- Yang Y, Zhang G, Hu C, et al. The germline mutational landscape of genitourinary cancers and its indication for prognosis and risk. BMC Urol 2022;22:196. [Crossref] [PubMed]
- Gordon MS, Hussey M, Nagle RB, et al. Phase II study of erlotinib in patients with locally advanced or metastatic papillary histology renal cell cancer: SWOG S0317. J Clin Oncol 2009;27:5788-93. [Crossref] [PubMed]
- Choueiri TK, Vaishampayan U, Rosenberg JE, et al. Phase II and biomarker study of the dual MET/VEGFR2 inhibitor foretinib in patients with papillary renal cell carcinoma. J Clin Oncol 2013;31:181-6. [Crossref] [PubMed]
- Li J, Jiang W, Wei J, et al. Patient specific circulating tumor DNA fingerprints to monitor treatment response across multiple tumors. J Transl Med 2020;18:293. [Crossref] [PubMed]
- Nalepa G, Clapp DW. Fanconi anaemia and cancer: an intricate relationship. Nat Rev Cancer 2018;18:168-85. [Crossref] [PubMed]
- Syed A, Tainer JA. The MRE11-RAD50-NBS1 Complex Conducts the Orchestration of Damage Signaling and Outcomes to Stress in DNA Replication and Repair. Annu Rev Biochem 2018;87:263-94. [Crossref] [PubMed]
- Ha GH, Ji JH, Chae S, et al. Pellino1 regulates reversible ATM activation via NBS1 ubiquitination at DNA double-strand breaks. Nat Commun 2019;10:1577. [Crossref] [PubMed]
- McDonnell KJ, Chemler JA, Bartels PL, et al. A human MUTYH variant linking colonic polyposis to redox degradation of the [4Fe4S]2+ cluster. Nat Chem 2018;10:873-80. [Crossref] [PubMed]
- Landrum MJ, Lee JM, Benson M, et al. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res 2018;46:D1062-7. [Crossref] [PubMed]
- Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 2015;17:405-24. [Crossref] [PubMed]
- Huang J, Cai W, Cai B, et al. Comprehensive Genomic Landscape in Chinese Clear Cell Renal Cell Carcinoma Patients. Front Oncol 2021;11:697219. [Crossref] [PubMed]
- Ho TH, Jonasch E. Genetic kidney cancer syndromes. J Natl Compr Canc Netw 2014;12:1347-55. [Crossref] [PubMed]
- van den Berg A, Buys CH. Involvement of multiple loci on chromosome 3 in renal cell cancer development. Genes Chromosomes Cancer 1997;19:59-76. [Crossref] [PubMed]
- Mehra R, Vats P, Cao X, et al. Somatic Bi-allelic Loss of TSC Genes in Eosinophilic Solid and Cystic Renal Cell Carcinoma. Eur Urol 2018;74:483-6. [Crossref] [PubMed]
- Yoo A, Tang C, Zucker M, et al. Genomic and Metabolic Hallmarks of SDH- and FH-deficient Renal Cell Carcinomas. Eur Urol Focus 2022;8:1278-88. [Crossref] [PubMed]
- Ricketts C, Woodward ER, Killick P, et al. Germline SDHB mutations and familial renal cell carcinoma. J Natl Cancer Inst 2008;100:1260-2. [Crossref] [PubMed]
- Lindor NM, Dechet CB, Greene MH, et al. Papillary renal cell carcinoma: analysis of germline mutations in the MET proto-oncogene in a clinic-based population. Genet Test 2001;5:101-6. [Crossref] [PubMed]
- Woodward ER, Clifford SC, Astuti D, et al. Familial clear cell renal cell carcinoma (FCRC): clinical features and mutation analysis of the VHL, MET, and CUL2 candidate genes. J Med Genet 2000;37:348-53. [Crossref] [PubMed]
- Xu Y, Kong W, Cao M, et al. Genomic Profiling and Response to Immune Checkpoint Inhibition plus Tyrosine Kinase Inhibition in FH-Deficient Renal Cell Carcinoma. Eur Urol 2023;83:163-72. [Crossref] [PubMed]
- Santos M, Lanillos J, Roldan-Romero JM, et al. Prevalence of pathogenic germline variants in patients with metastatic renal cell carcinoma. Genet Med 2021;23:698-704. [Crossref] [PubMed]
- McLaughlin CN, Perry-Richardson JJ, Coutinho-Budd JC, et al. Dying Neurons Utilize Innate Immune Signaling to Prime Glia for Phagocytosis during Development. Dev Cell 2019;48:506-22.e6. [Crossref] [PubMed]
- Tang T, Tan X, Wang Z, et al. Germline Mutations in Patients With Early-Onset Prostate Cancer. Front Oncol 2022;12:826778. [Crossref] [PubMed]
- Linehan WM. Genetic basis of kidney cancer: role of genomics for the development of disease-based therapeutics. Genome Res 2012;22:2089-100. [Crossref] [PubMed]
- Mullin BH, Tickner J, Zhu K, et al. Characterisation of genetic regulatory effects for osteoporosis risk variants in human osteoclasts. Genome Biol 2020;21:80. [Crossref] [PubMed]
- Ricketts CJ, Shuch B, Vocke CD, et al. Succinate dehydrogenase kidney cancer: an aggressive example of the Warburg effect in cancer. J Urol 2012;188:2063-71. [Crossref] [PubMed]
- Giráldez MD, Balaguer F, Bujanda L, et al. MSH6 and MUTYH deficiency is a frequent event in early-onset colorectal cancer. Clin Cancer Res 2010;16:5402-13. [Crossref] [PubMed]
- Magrin L, Fanale D, Brando C, et al. MUTYH-associated tumor syndrome: The other face of MAP. Oncogene 2022;41:2531-9. [Crossref] [PubMed]
- Bogliolo M, Bluteau D, Lespinasse J, et al. Biallelic truncating FANCM mutations cause early-onset cancer but not Fanconi anemia. Genet Med 2018;20:458-63. [Crossref] [PubMed]
- Tan W, Deans AJ. The ubiquitination machinery of the Fanconi Anemia DNA repair pathway. Prog Biophys Mol Biol 2021;163:5-13. [Crossref] [PubMed]
- Brandt S, Samartzis EP, Zimmermann AK, et al. Lack of MRE11-RAD50-NBS1 (MRN) complex detection occurs frequently in low-grade epithelial ovarian cancer. BMC Cancer 2017;17:44. [Crossref] [PubMed]
- Bian L, Meng Y, Zhang M, et al. MRE11-RAD50-NBS1 complex alterations and DNA damage response: implications for cancer treatment. Mol Cancer 2019;18:169. [Crossref] [PubMed]
- Chen M, Linstra R, van Vugt MATM. Genomic instability, inflammatory signaling and response to cancer immunotherapy. Biochim Biophys Acta Rev Cancer 2022;1877:188661. [Crossref] [PubMed]
- Che R, Zhang J, Nepal M, et al. Multifaceted Fanconi Anemia Signaling. Trends Genet 2018;34:171-83. [Crossref] [PubMed]
- Webster ALH, Sanders MA, Patel K, et al. Genomic signature of Fanconi anaemia DNA repair pathway deficiency in cancer. Nature 2022;612:495-502. [Crossref] [PubMed]
- Berwick M, Satagopan JM, Ben-Porat L, et al. Genetic heterogeneity among Fanconi anemia heterozygotes and risk of cancer. Cancer Res 2007;67:9591-6. [Crossref] [PubMed]
- McCarthy-Leo C, Darwiche F, Tainsky MA. DNA Repair Mechanisms, Protein Interactions and Therapeutic Targeting of the MRN Complex. Cancers (Basel) 2022;14:5278. [Crossref] [PubMed]
- Theunissen JW, Kaplan MI, Hunt PA, et al. Checkpoint failure and chromosomal instability without lymphomagenesis in Mre11(ATLD1/ATLD1) mice. Mol Cell 2003;12:1511-23. [Crossref] [PubMed]
- Gupta GP, Vanness K, Barlas A, et al. The Mre11 complex suppresses oncogene-driven breast tumorigenesis and metastasis. Mol Cell 2013;52:353-65. [Crossref] [PubMed]
- Williams BR, Mirzoeva OK, Morgan WF, et al. A murine model of Nijmegen breakage syndrome. Curr Biol 2002;12:648-53. [Crossref] [PubMed]
- Kang J, Ferguson D, Song H, et al. Functional interaction of H2AX, NBS1, and p53 in ATM-dependent DNA damage responses and tumor suppression. Mol Cell Biol 2005;25:661-70. [Crossref] [PubMed]
- Buis J, Wu Y, Deng Y, et al. Mre11 nuclease activity has essential roles in DNA repair and genomic stability distinct from ATM activation. Cell 2008;135:85-96. [Crossref] [PubMed]
- Luo G, Yao MS, Bender CF, et al. Disruption of mRad50 causes embryonic stem cell lethality, abnormal embryonic development, and sensitivity to ionizing radiation. Proc Natl Acad Sci U S A 1999;96:7376-81. [Crossref] [PubMed]
- Zhu J, Petersen S, Tessarollo L, et al. Targeted disruption of the Nijmegen breakage syndrome gene NBS1 leads to early embryonic lethality in mice. Curr Biol 2001;11:105-9. [Crossref] [PubMed]
- Smogorzewska A, Matsuoka S, Vinciguerra P, et al. Identification of the FANCI protein, a monoubiquitinated FANCD2 paralog required for DNA repair. Cell 2007;129:289-301. [Crossref] [PubMed]
- Rohleder F, Huang J, Xue Y, et al. FANCM interacts with PCNA to promote replication traverse of DNA interstrand crosslinks. Nucleic Acids Res 2016;44:3219-32. [Crossref] [PubMed]
- Liang Y, Chiu PK, Zhu Y, et al. Whole-exome sequencing reveals a comprehensive germline mutation landscape and identifies twelve novel predisposition genes in Chinese prostate cancer patients. PLoS Genet 2022;18:e1010373. [Crossref] [PubMed]
- Alter BP, Rosenberg PS. VACTERL-H Association and Fanconi Anemia. Mol Syndromol 2013;4:87-93. [Crossref] [PubMed]
- Neveling K, Endt D, Hoehn H, et al. Genotype-phenotype correlations in Fanconi anemia. Mutat Res 2009;668:73-91. [Crossref] [PubMed]
- Nie D, Zhang J, Wang F, et al. Fanconi anemia gene-associated germline predisposition in aplastic anemia and hematologic malignancies. Front Med 2022;16:459-66. [Crossref] [PubMed]
- Kwong A, Ho CYS, Shin VY, et al. Germline mutations in Chinese ovarian cancer with or without breast cancer. Mol Genet Genomic Med 2022;10:e1940. [Crossref] [PubMed]
- Zdzienicka MZ, Arwert F. Breast cancer and Fanconi anemia: what are the connections? Trends Mol Med 2002;8:458-60. [Crossref] [PubMed]
- Seal S, Barfoot R, Jayatilake H, et al. Evaluation of Fanconi Anemia genes in familial breast cancer predisposition. Cancer Res 2003;63:8596-9. [PubMed]
- García MJ, Fernández V, Osorio A, et al. Mutational analysis of FANCL, FANCM and the recently identified FANCI suggests that among the 13 known Fanconi Anemia genes, only FANCD1/BRCA2 plays a major role in high-risk breast cancer predisposition. Carcinogenesis 2009;30:1898-902. [Crossref] [PubMed]
- Bakker JL, Thirthagiri E, van Mil SE, et al. A novel splice site mutation in the noncoding region of BRCA2: implications for Fanconi anemia and familial breast cancer diagnostics. Hum Mutat 2014;35:442-6. [Crossref] [PubMed]
- Kohlhase S, Bogdanova NV, Schürmann P, et al. Mutation analysis of the ERCC4/FANCQ gene in hereditary breast cancer. PLoS One 2014;9:e85334. [Crossref] [PubMed]
- Abbasi S, Rasouli M. A rare FANCA gene variation as a breast cancer susceptibility allele in an Iranian population. Mol Med Rep 2017;15:3983-8. [Crossref] [PubMed]
- Rogers CD, van der Heijden MS, Brune K, et al. The genetics of FANCC and FANCG in familial pancreatic cancer. Cancer Biol Ther 2004;3:167-9. [Crossref] [PubMed]
- Esteban-Jurado C, Franch-Expósito S, Muñoz J, et al. The Fanconi anemia DNA damage repair pathway in the spotlight for germline predisposition to colorectal cancer. Eur J Hum Genet 2016;24:1501-5. [Crossref] [PubMed]
- Wagner JE, Tolar J, Levran O, et al. Germline mutations in BRCA2: shared genetic susceptibility to breast cancer, early onset leukemia, and Fanconi anemia. Blood 2004;103:3226-9. [Crossref] [PubMed]
- Wang L, Wang H, Wang T, et al. Analysis of polymorphisms in genes associated with the FA/BRCA pathway in three patients with multiple primary malignant neoplasms. Artif Cells Nanomed Biotechnol 2019;47:1101-12. [Crossref] [PubMed]
- Savage SA, Ballew BJ, Giri N, et al. Novel FANCI mutations in Fanconi anemia with VACTERL association. Am J Med Genet A 2016;170A:386-91. [Crossref] [PubMed]
(English Language Editor: J. Jones)