
CENPF promotes the proliferation of renal cell carcinoma in vitro
Highlight box
Key findings
• This study found that CENPF was highly expressed in RCC tissues. Moreover, down-regulation of CENPF can significantly inhibit the proliferation of RCC cells and change the distribution of cell cycle. CENPF plays a tumorigenic role in RCC.
What is known and what is new?
• CENPF, a microtubule binding protein, has been highlighted the correlation with cancer progression in various types of cancer, which is significantly correlated with OS and RFS in clear cell RCC;
• Our new finding is that knockdown CENPF can significantly inhibit the proliferation of RCC cells and change the distribution of cell cycle.
What is the implication, and what should change now?
• CENPF is a potential oncogene and prognostic marker in RCC, which can be used as an independent prognostic factor.
Introduction
Renal cell carcinoma (RCC) is the ninth most common cancer worldwide. It is estimated that there were more than 337,000 new cases diagnosed in 2012. An estimated 121,000 new cases of RCC were diagnosed in 2012 in Europe, of which more than 75,000 affected men (1). Clear cell renal carcinoma is the most common type of RCC. Although the prognosis of RCC patients has been remarkably improved due to nephrectomy, ablative therapies, and targeted therapies, a large proportion of stage III and IV disease still leads to mortality and lower quality of life. Therefore, it is necessary to develop new targeted therapies against RCC.
Centromere protein F (CENPF), a microtubule binding protein, is a component of the nuclear matrix during the G2 phase of the cell cycle. It localizes to the spindle midzone and the intracellular bridge in late anaphase and telophase, respectively, and CENPF mutation may lead to Stromme syndrome (2). A recent study showed that CENPF also serves as a centriolar disease-related gene implicated in severe human ciliopathy, namely, Stromme syndrome, and microcephaly-related phenotypes (3). In mouse embryos, CENPF depletion causes developmental failure (4).
Notably, a study has reported the function and dysregulation of CENPF in various types of cancer. For instance, reduced CENPF remodels prostate cancer cells and regulates cellular metabolism (5). CENPF contributes to prostate cancer aggressiveness, and its interaction with FOXM1 drives prostate cancer malignancy (6,7). Another study found that CENPF regulated cancer metabolism by pyruvate kinase M2 phosphorylation signaling (8). Among hepatocellular carcinoma (HCC) patients, CENPF is one of the frequently amplified genes (9). The overexpression of CENPF has been shown to predict shorter overall survival (OS) and higher cumulative recurrence in HCC and breast cancer (10-12). Taken together, CENPF has been shown to promote proliferation and metastasis in different types of cancer and to predict poor prognosis.
However, the functions of CENPF in RCC remain elusive. Microarray dataset analysis has indicated that MT2A, MYC, CENPF, and NEK2 have a high degree of participation in RCC (13). The expression level of CENPF has been significantly correlated with OS and recurrence-free survival (RFS) in clear cell RCC (14,15). In this study, we aimed to elucidate the clinical significance and potential roles of CENPF in RCC. We present the following article in accordance with the MDAR reporting checklist (available at https://tau.amegroups.com/article/view/10.21037/tau-22-797/rc).
Methods
Quantitative real-time polymerase chain reaction (qRT-PCR)
Peripheral blood and corresponding tissue samples of 23 RCC patients and 23 normal physical examination patients who were treated in our hospital from 2018 to 2020 were collected. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the Institutional Review Board of Nantong First People’s Hospital (No. LI2017-059) and informed consent was taken from all individual participants. Total RNA was extracted with an RNeasy® kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. A total of 500 ng RNA was used to generate complementary DNA (cDNA) for each sample. The polymerase chain reaction (PCR) was preceded by 94 ℃ for 5 minutes, followed by 30 cycles of 94 ℃ for 45 seconds, 55 ℃ for 45 seconds, and 72 ℃ for 1 minute followed by 72 ℃ for 7 minutes. The cycle threshold (Ct) values were normalized to the expression levels of glyceraldehyde 3-phosphate dehydrogenase (GAPDH), and the relative amount of mRNA specific to each of the target genes was calculated via the 2−ΔΔCt method. The sequences of primers are as follows:
- CENPF-F: AAACAAGATTCCCGAGGGTCTC;
- CENPF-R: GCCTGAAGCTTTATTTTGGCCA;
- CDK4-F: TGCTGGATGTCATTCACACAGA;
- CDK4-R: TTGATGAGGGGAAGAGGAATGC;
- CDK6-F: GCCTTGCCCGCATCTATAGT;
- CDK6-R: GCAGCCAACACTCCAGAGAT;
- CyclinD1-F: TTCGTGGCCTCTAAGATGAAGG;
- CyclinD1-R: GTTCCACTTGAGCTTGTTCACC;
- GAPDH: TGGTCTCCTCTGACTTCAACAG
- GAPDH-R: CCCTGTTGCTGTAGCCAAATTC.
Cell culture and siRNA transfection
The HK2, 786-O, ACHN, and A498 cell lines were cultured in Roswell Park Memorial Institute (RPMI)-1640 supplemented with 10% fetal bovine serum (FBS; Gibco, Waltham, MA, USA), 100 U/mL penicillin, and 100 µg/mL streptomycin at 37 ℃ in the presence of 5% CO2. siCENPF was transfected into ACHN and A498 cells by using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol. The siRNA sequences are as follows:
- siCENPF-1: GGGUUCUCUUACCCUGAGAAUGA;
- siCENPF-2: CGUCGUACUUAAUGUCUGUUAGA.
Protein extraction and Western blot analysis
Cell lysates were extracted with mammalian protein extraction reagent (M-PER) lysis buffer supplemented with a protease inhibitor and phosphatase inhibitors. The protein concentrations were measured by a Pierce bicinchoninic acid (BCA) protein assay kit (Thermo Fisher, Waltham, MA, USA). A total of 30 µg of protein was loaded onto a sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) gel and run with 1× SDS running buffer. Proteins were transferred onto a 0.45 µm polyvinylidene fluoride (PVDF) membrane, which was later blocked with 5% milk in tris-buffered saline (TBS)-0.1% Tween-20. Then, the membrane was incubated with anti-CENPF (1:1,000; Proteintech, Rosemont, IL, USA) or anti-β-actin (1:5,000; Proteintech) primary antibody overnight at 4 ℃ and secondary antibody for 1 hour. Finally, proteins were detected using enhanced chemiluminescence (ECL).
Immunohistochemistry (IHC)
Formalin-fixed paraffin-embedded (FFPE) tissue samples were cut into 4 µm thick sections and dried at 60 ℃ for 30 minutes. After blocking endogenous peroxidase activity using blocking reagent (Dako, Santa Clara, CA, USA) for 5 minutes at room temperature, epitopes were retrieved using pH 9.0 Tris-ethylenediamine tetraacetic acid (EDTA) buffer for 40 minutes at 95 ℃ followed by incubation with anti-CENPF (1:200, Proteintech) antibody at room temperature for 30 minutes. Subsequently, the slides were incubated with peroxidase/diaminobenzidine (DAB)-10 min K5007 from a DAKO EnVisionTM Detection Kit (Dako) for 30 minutes.
Proliferation assay
In the Cell Counting Kit-8 (CCK-8) assay, ACHN and A498 cells were plated in 96-well plates at a density of 2,000 cells/well. On days 1, 2, 3, and 4, 10 µL CCK-8 was added to each well. After incubation for 3 hours, the absorbance [optical density (OD) value] was measured at 450 nm. In the clonogenic assay, ACHN and A498 cells were plated in 6-well plates at a density of 400 cells/well. On day 7, the clones were stained with crystal violet.
Immunofluorescence
The cells were seeded on coverslips in 6-well plates. After siRNA transfection, cells were fixed in 4% paraformaldehyde for 30 minutes and then blocked with 0.3% bovine serum albumin (BSA) and 0.3% Triton X-100 to block nonspecific binding. The coverslips were incubated with a primary antibody against EdU (RiboBio, Guangzhou, China) at 4 ℃ overnight, followed by a secondary antibody against Apollo488 for 1 hour at room temperature. Images were captured under a fluorescence microscope.
Cell cycle analysis
Cell cycle was analyzed by flow cytometry. Briefly, after fixation, the cells were prepared for propidium iodide (PI; Sigma, St. Louis, MO, USA) staining according to the manufacturer’s protocol. DNA content was determined using a FACSCaliber Analyzer [Becton, Dickinson, and Co. (BD), Franklin Lakes, NJ, USA] and analyzed by FlowJo (BD, USA).
Statistical analysis
All data were analyzed by SPSS 19.0 (IBM Corp., Chicago, IL, USA). Statistical analysis of significance was performed by one-way analysis of variance (ANOVA) with least significant difference (LSD) post-hoc multiple comparisons. Student’s t-test was used to compare the differences between 2 groups. A P value <0.05 indicated significance.
Results
CENPF was highly expressed in RCC and predicted poor prognosis
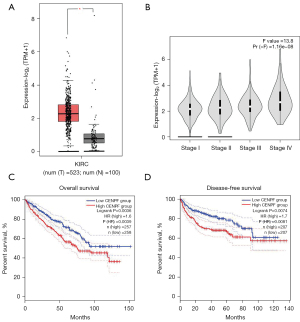
We downloaded and analyzed the RNA sequencing and clinical data from The Cancer Genome Atlas (TCGA) cancer database (https://portal.gdc.cancer.gov). Our results showed that CENPF was highly expressed in kidney renal clear cell carcinoma (KIRC) compared to normal kidney tissue (P<0.05, Figure 1A). Notably, stage IV tumors expressed higher CENPF than stage I and II tumors (Figure 1B). In addition, higher CENPF expression predicted poorer OS [P=0.0036, hazard ratio (HR) =1.6] and disease-free survival (DFS) (P=0.0074, HR =1.7, Figure 1C,1D). These results suggested that CENPF may play a role in RCC progression.
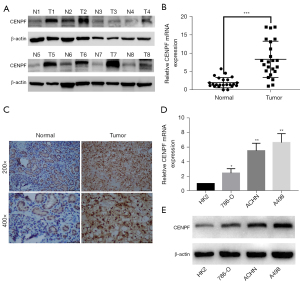
Moreover, we validated CENPF expression in our cell line and cancer tissue. We collected 23 paired RCC tissues and paracancerous kidney tissues. We assessed CENPF expression by quantitative polymerase chain reaction (qPCR), Western blotting, and IHC. The results confirmed that CENPF was highly expressed in tumor samples compared to normal tissue at both the messenger RNA (mRNA) and protein levels (Figure 2A-2C). Among the 4 RCC cell lines we evaluated (HK2, 786-O, ACHN, and A498), HK2 showed the lowest CENPF expression level, whereas A498 showed the highest (Figure 2D,2E).
Knockdown of CENPF inhibited the proliferation of RCC cells
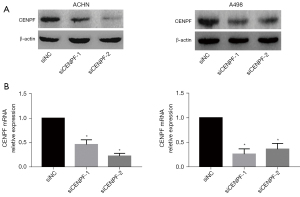
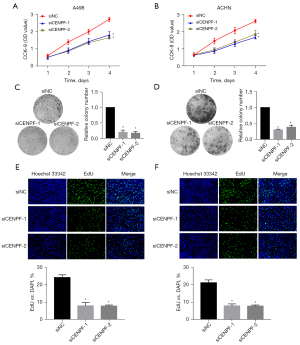
We further studied the roles of CENPF in RCC cell lines in vitro. We downregulated CENPF in ACHN and A498 cells via small interfering RNA (siRNA). Both siRNAs effectively reduced CENPF mRNA and protein (Figure 3). The CCK-8 assay and clonogenic assay revealed that reduced CENPF significantly inhibited RCC proliferation in vitro (Figure 4A,4B). Inhibition of CENPF reduced clone formation in vitro (Figure 4C,4D). We also performed 5-ethynyl-2'-deoxyuridine (EdU) staining by immunofluorescence. CENPF knockdown significantly reduced the EdU signal, which is a marker of cell proliferation (Figure 4E,4F). These results suggested that CENPF might contribute to the proliferation of RCC.
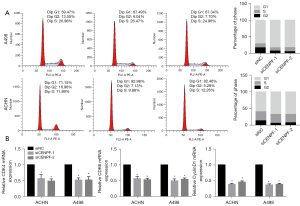
Inhibition of CENPF regulated cell cycle distribution in RCC cells
Since CENPF participates in chromosome segregation during mitosis, we investigated whether CENPF regulated the cell cycle in RCC. Our results indicated that CENPF-depleted cells exhibited a lower proportion in G1 phase but a higher proportion in G2 phase in ACHN and A498 cells (Figure 5A). This suggested that CENPF inhibition promoted cells to enter G1 phase. Furthermore, we studied the mRNA expression of cell cycle regulators in G1 phase. CENPF inhibition by siRNA significantly reduced the mRNA levels of CDK4, CDK6, and CyclinD1 (P<0.05, Figure 5B). These results confirmed that CENPF promoted RCC proliferation via cell cycle regulation in vitro.
Discussion
Patients with RCC recurrence or metastasis show poor prognosis. There are already several potential new biomarkers of RCC progression and treatment (16-18). Higher inflammatory indicators such as vascular endothelial growth factor (VEGF) were found in the tumor microenvironment of RCC, VEGF pathway inhibitors and mammalian target of rapamycin (mTOR) inhibitors are the main targeted therapies against RCC. However, the prognosis of advanced RCC patients is still limited if the tumors become resistant to these therapies. Cell mitosis is, to some extent, dependent on the centromere-kinetochore complex, especially cancer cells. Therefore, kinetochore activity may be a promising drug target for cancer treatment. CENPF plays important roles in the cell cycle and division. During mitosis, ataxia telangiectasia mutated and Rad3-related (ATR) kinase localizes to centromeres through Aurora A-regulated association with CENPF, allowing ATR to engage replication protein A (RPA)-coated centromeric R loops. After activation, ATR then stimulates Aurora B, preventing lagging chromosomes (19). In our study, we reported the protumor activity of CENPF in RCC cell lines for the first time. It provides a basis for the development of targeted drugs in the future.
Our results may provide new insights into RCC proliferation and malignancy. In this study, we revealed that CENPF is highly expressed in RCC tissue and that downregulation of CENPF can significantly inhibit RCC cell proliferation and change cell cycle distribution. These results are consistent with previous studies (13,14). Interestingly, many studies have reported that several genes and noncoding RNAs (ncRNAs) can promote cancer cell proliferation or metastasis by modulating CENPF. For instance, lncRNA MCM3AP-antisense 1 (MCM3AP-AS1) contributes to breast cancer cell progression through the miR-28-5p/CENPF axis (20). Chen et al. reported that the HnRNPR-CCNB1/CENPF axis promotes gastric cancer proliferation and metastasis (21). This evidence may further confirm the protumor activity of CENPF. However, how CENPF is overexpressed in RCC cells is still unclear.
Cell cycle progression is one of the determinants of cancer cell malignancy. Cancer cells need to pass from the G1 phase into S phase through a tightly regulated checkpoint. Specifically, the G1/S transition begins in early G1, triggering an increase in D-type cyclins (D1, D2, and D3), which bind to CDK4 or CDK6. These cyclin-CDK complexes translocate to the nucleus, where they are phosphorylated. In turn, activated CDK4/6 complexes phosphorylate retinoblastoma (RB) tumor suppressor proteins. RB reduces the expression of S phase genes by directly inhibiting E2F transactivation (22). Therefore, the G1/S transition largely depends on the activity of CDK4/6 and D-type cyclins (23). In our study, we found that CENPF inhibition significantly reduced CDK4/6 and CyclinD1 expression (24), which may partially explain the phenotype. Our results provide evidence that CENPF downregulation or mutation may sensitize RCC tumors to cell cycle inhibitors.
In summary, this project found that the expression of CENPF is upregulated in RCC and that downregulation of CENPF can inhibit the proliferation of RCC cells. It also clarified the mechanism by which CENPF regulates the cell cycle of RCC by inhibiting the expression of cyclins such as CDK4, CDK6, and CyclinD1. This project has discovered new pathogenic genes and pathogenesis in RCC, which provides a scientific basis for exploring new therapeutic targets.
Conclusions
CENPF can be used as an independent prognostic factor for RCC. Down-regulation of CENPF can significantly inhibit the proliferation of RCC cells and change the distribution of cell cycle. CENPF is a potential oncogene and prognostic marker in RCC.
Acknowledgments
Funding: This work was supported by the Nantong Science and Technology Department Projects (Nos. MS22022085, MS22019009, JCZ2022075, and JC12022008).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://tau.amegroups.com/article/view/10.21037/tau-22-797/rc
Data Sharing Statement: Available at https://tau.amegroups.com/article/view/10.21037/tau-22-797/dss
Peer Review File: Available at https://tau.amegroups.com/article/view/10.21037/tau-22-797/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tau.amegroups.com/article/view/10.21037/tau-22-797/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the Institutional Review Board of Nantong First People’s Hospital (No. LI2017-059) and informed consent was taken from all individual participants.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Jonasch E, Gao J, Rathmell WK. Renal cell carcinoma. BMJ 2014;349:g4797. [Crossref] [PubMed]
- Ozkinay F, Atik T, Isik E, et al. A further family of Stromme syndrome carrying CENPF mutation. Am J Med Genet A 2017;173:1668-72. [Crossref] [PubMed]
- Waters AM, Asfahani R, Carroll P, et al. The kinetochore protein, CENPF, is mutated in human ciliopathy and microcephaly phenotypes. J Med Genet 2015;52:147-56. [Crossref] [PubMed]
- Zhou CJ, Wang XY, Han Z, et al. Loss of CENPF leads to developmental failure in mouse embryos. Cell Cycle 2019;18:2784-99. [Crossref] [PubMed]
- Shahid M, Kim M, Lee MY, et al. Downregulation of CENPF Remodels Prostate Cancer Cells and Alters Cellular Metabolism. Proteomics 2019;19:e1900038. [Crossref] [PubMed]
- Aytes A, Mitrofanova A, Lefebvre C, et al. Cross-species regulatory network analysis identifies a synergistic interaction between FOXM1 and CENPF that drives prostate cancer malignancy. Cancer Cell 2014;25:638-51. [Crossref] [PubMed]
- Göbel C, Özden C, Schroeder C, et al. Upregulation of centromere protein F is linked to aggressive prostate cancers. Cancer Manag Res 2018;10:5491-504. [Crossref] [PubMed]
- Shahid M, Lee MY, Piplani H, et al. Centromere protein F (CENPF), a microtubule binding protein, modulates cancer metabolism by regulating pyruvate kinase M2 phosphorylation signaling. Cell Cycle 2018;17:2802-18. [Crossref] [PubMed]
- Kim HE, Kim DG, Lee KJ, et al. Frequent amplification of CENPF, GMNN and CDK13 genes in hepatocellular carcinomas. PLoS One 2012;7:e43223. [Crossref] [PubMed]
- Yang X, Miao BS, Wei CY, et al. Lymphoid-specific helicase promotes the growth and invasion of hepatocellular carcinoma by transcriptional regulation of centromere protein F expression. Cancer Sci 2019;110:2133-44. [Crossref] [PubMed]
- Zhuo YJ, Xi M, Wan YP, et al. Enhanced expression of centromere protein F predicts clinical progression and prognosis in patients with prostate cancer. Int J Mol Med 2015;35:966-72. [Crossref] [PubMed]
- Sun J, Huang J, Lan J, et al. Overexpression of CENPF correlates with poor prognosis and tumor bone metastasis in breast cancer. Cancer Cell Int 2019;19:264. [Crossref] [PubMed]
- Cheng Y, Hong M, Cheng B. Identified differently expressed genes in renal cell carcinoma by using multiple microarray datasets running head: differently expressed genes in renal cell carcinoma. Eur Rev Med Pharmacol Sci 2014;18:1033-40. [PubMed]
- Wei W, Lv Y, Gan Z, et al. Identification of key genes involved in the metastasis of clear cell renal cell carcinoma. Oncol Lett 2019;17:4321-8. [Crossref] [PubMed]
- Yamada Y, Arai T, Kojima S, et al. Regulation of antitumor miR-144-5p targets oncogenes: Direct regulation of syndecan-3 and its clinical significance. Cancer Sci 2018;109:2919-36. [Crossref] [PubMed]
- Peng X, Pan X, Liu K, et al. miR-142-3p as a novel biomarker for predicting poor prognosis in renal cell carcinoma patients after surgery. Int J Biol Markers 2019;34:302-8. [Crossref] [PubMed]
- Ellinger J, Poss M, Brüggemann M, et al. Systematic Expression Analysis of Mitochondrial Complex I Identifies NDUFS1 as a Biomarker in Clear-Cell Renal-Cell Carcinoma. Clin Genitourin Cancer 2017;15:e551-62. [Crossref] [PubMed]
- Li P, Liu J, Li J, et al. DNA methylation of CRB3 is a prognostic biomarker in clear cell renal cell carcinoma. Mol Biol Rep 2019;46:4377-83. [Crossref] [PubMed]
- Kabeche L, Nguyen HD, Buisson R, et al. A mitosis-specific and R loop-driven ATR pathway promotes faithful chromosome segregation. Science 2018;359:108-14. [Crossref] [PubMed]
- Chen Q, Xu H, Zhu J, et al. LncRNA MCM3AP-AS1 promotes breast cancer progression via modulating miR-28-5p/CENPF axis. Biomed Pharmacother 2020;128:110289. [Crossref] [PubMed]
- Chen EB, Qin X, Peng K, et al. HnRNPR-CCNB1/CENPF axis contributes to gastric cancer proliferation and metastasis. Aging (Albany NY) 2019;11:7473-91. [Crossref] [PubMed]
- Goel S, DeCristo MJ, McAllister SS, et al. CDK4/6 Inhibition in Cancer: Beyond Cell Cycle Arrest. Trends Cell Biol 2018;28:911-25. [Crossref] [PubMed]
- Li P, Hao Z, Zeng F. Tumor suppressor stars in yeast G1/S transition. Curr Genet 2021;67:207-12. [Crossref] [PubMed]
- Quan LL, Liu JY, Qu LX, et al. Expression of Cyclin D1 gene in ovarian cancer and effect of silencing its expression on ovarian cancer cells based on the Oncomine database. Bioengineered 2021;12:9290-300. [Crossref] [PubMed]
(English Language Editor: J. Jones)


