
KIF11 is a potential prognostic biomarker and therapeutic target for adrenocortical carcinoma
Highlight box
Key findings
• KIF11 could be a predictor of poor prognosis and possibly serve as a novel therapeutic target for ACC
What is known and what is new?
• ACC is a rare and aggressive malignant endocrine neoplasia of the adrenal cortex, ACC prognosis remains poor, and novel treatments and prognostic markers are urgently required.
• High KIF11 expression levels are significantly associated with the progression of ACC and resulting in poor survival rates. KIF11 expression appears to be a novel and independent prognostic marker in ACC. KIF11 is a key protein involved in cell proliferation and invasion, and may thus serve as a potential novel therapeutic target for ACC.
What is the implication, and what should change now?
• Treatment options are limited for patients with advanced ACC.
Introduction
Adrenocortical carcinoma (ACC) is a rare and aggressive malignant endocrine neoplasia of the adrenal cortex with an incidence rate of 0.5 to 2 cases per million population per year (1). For patients with localized ACC, complete operative resection is the preferred treatment for long-term survival (2). Unfortunately, ACC is a highly malignant tumor, with 70% to 85% of patients experiencing recurrence after surgical resection (3,4). Initial staging of ACC is the most important factor in predicting its prognosis. The 5-year overall survival (OS) rate ranges from 60% to 80% in patients with stage I ACC and decreases to 13% in those with stage IV ACC (5).
Although several factors have been reported to be associated with the prognosis and risk of tumor recurrence, an accurate prediction of patients with ACC remains a challenge. Moreover, treatment options are limited for patients with advanced ACC. Currently, the major of adjuvant treatment consists of mitotane alone or in combination with multi-drug chemotherapeutics, such as etoposide, doxorubicin, and cisplatin, which is known as the Italian protocol (6). However, this multi-drug treatment regimen has significant toxicity potential in patients. Furthermore, mitotane, which has a specific cytotoxic effect on the steroidogenic cells of the adrenal cortex, is the only approved drug for the treatment of advanced ACC (7,8). The side effects of mitotane treatment include toxicity affecting the bone marrow, liver, skin, gastrointestinal tract, and neuromuscular junctions (8). Therefore, ACC prognosis remains poor, and novel treatments and prognostic markers are urgently required.
Kinesin family member 11 (KIF11), also known as EG5 or kinesin-5, is thought to be vital in the tetrameric microtubule cross-linkage, cell mitosis, cell cycle, and cell differentiation (9,10). Although the physiological function of KIF11 remains largely unclear, studies have suggested that high KIF11 expression levels are associated with advanced stages of cancer, invasion, metastasis, and recurrence. Recently, KIF11 is shown to be overexpressed in several tumors and associated with poor prognosis of diseases such as breast cancer (11), liver cancer (12), prostate cancer (13), bladder cancer (14), clear cell renal cell carcinoma (15), gastric cancer (16), non-small cell lung cancer (17), and meningioma (18). However, the relevant roles and mechanisms of KIF11 in ACC progression have not been studied yet. Monastrol, a cytotoxic small molecule, from dihydropyrimidinone scaffold, is an inhibitor of KIF11. The effect of Monastrol on ACC is yet to be verified. This study aimed to evaluate the clinical significance and therapeutic potential of KIF11 protein in ACC. We present the following article in accordance with the STROBE reporting checklist (available at https://tau.amegroups.com/article/view/10.21037/tau-22-706/rc).
Methods
Differential expression of KIF11: The Cancer Genome Atlas (TCGA) and Genotype Tissue Expression (GTEx) database analyses
TCGA database (n=79) and GTEx database (n=128) were utilized to explore the differential expression of KIF11 in ACC and normal adrenal tissues. The data is RNA-seq data in transcripts per million reads (TPM) format, which is uniformly processed by Toil process (19). KIF11 differential expression in the ACC tissues in the TCGA database was defined as KIF11-high or KIF11-low when the values were above or below the KIF11 median value, respectively. Detailed clinical information, including sex, age, TNM stages, pathologic stages, therapy outcome, and survival information, including the OS, disease-specific survival (DSS), and progress-free interval (PFI), for the ACC samples was obtained from the TCGA.
Differentially expressed genes (DEGs) analysis, correlation analysis, Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis, and Gene set enrichment analysis (GSEA)
DEGs in patients with KIF11-high and KIF11-low expression in the TCGA dataset were identified using the unpaired Student’s t-test and DESeq2 (1.26.0) package (20). Genes with an adjusted P value <0.05, and log2 fold change >2.0, were considered statistically significant. GO and KEGG pathway enrichment analyses were performed using the clusterProfiler package in R (21). Adjusted P values <0.05 were considered to indicate statistically significant pathways. GSEA was performed using the GSEA software (22).
Ethical statement
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of the Xiangya Hospital, China (No. 202109889). And all samples and clinical information used in this study were obtained with informed consent from patients.
ACC samples and normal tissues
ACC samples (n=30) and normal tissues (n=20) were collected from patients undergoing surgery at Xiangya Hospital between January 2009 and January 2016. The ACC inclusion criteria were as follows: (I) unilateral tumor, (II) postoperative pathologic diagnosis of ACC, (III) no other systemic diseases, (IV) and no other treatment before surgery. Hematoxylin and eosin (H&E)-stained sections were reviewed by two independent pathologists for confirmation. Clinicopathological parameters, such as age, sex, clinical symptoms, tumor size, stages, metastasis status, and survival data, were extracted from the medical records, pathological reports, and patient follow-up. Follow-up was done for all patients 72 months after surgery.
H&E staining
ACC and normal tissues were fixed in 4% paraformaldehyde solution and embedded in paraffin. For H&E staining, slides were immersed in a hematoxylin solution for 3 to 5 min, differentiated with acid alcohol, and counterstained with eosin for 3 min. Images of the H&E-stained sections were acquired using a microscope (Nikon, Japan).
Immunohistochemical (IHC) staining and analysis
For IHC staining, the paraffin sections were deparaffinized, rehydrated, immersed in 3% hydrogen peroxide for 10 min to inactivate endogenous peroxidase, and then incubated for 30 min in blocking buffer containing 5% bovine serum albumin. After blocking, the sections were incubated with primary antibodies against KIF11 (1:100; Proteintech, China) overnight at 4 ℃, followed by incubation at room temperature with horseradish peroxidase-conjugated anti-rabbit secondary antibody (1:100; Sigma-Aldrich, USA). The IHC staining results for the KIF11 proteins were determined using a semi-quantitative analysis technique, in which samples were analyzed using a bright-field microscope (Nikon, Japan). For each sample, the protein expression intensity was determined using ImageJ software. The KIF11 values were expressed as the percentage of positive cells in each case. The cell staining intensity was divided into two categories: cases with greater than or equal to 30% positive nuclei were classified as KIF11-high group, and those with less than 30% were classified as KIF11-low group. Three independent observers inspected the specimens in a blinded manner.
Cell culture and treatment
Human ACC cell lines, NCI-H295R (hormonally active) were purchased from Procell Life Science & Technology Co., Ltd. (Wuhan, China). NCI-H295R cell were cultured in DMEM (Procell, China) supplemented with 10% fetal bovine serum (FBS; CellMax, Australia) and 1% penicillin/streptomycin in a humidified 5% CO2 atmosphere at 37 ℃.
Cell viability assay
For the cell counting kit-8 (CCK-8) assay, cells (1×104 cells/well) were seeded into 96-well plates and cultured with or without mitotane or monastrol (3, 10, 30, and 100 µM) for 48 h. Subsequently, 10 µL of CCK-8 solution (Dojindo, Japan) was added to each well at different time points. After 2 h of incubation, the optical density at 450 nm was measured using a microplate reader (Thermo Scientific, USA).
Transwell assay
A cell invasion assay was performed using 8.0 µm PET Membrane 24-well Transwell chamber (Millipore, USA). For the invasion assay, the chamber was coated with Matrigel (BD Biosciences, USA), cells were seeded into the upper chamber with serum-free medium (2×105 cells), and DMEM with 20% FBS was added to the bottom chamber with or without mitotane or monastrol (30 µM) for 24 hours. After incubation for 24 hours, the cells were fixed in 4% paraformaldehyde and stained with 1% crystal violet. Cells that had migrated were counted under an inverted light microscope by counting the number of cells from 10 random fields at 100× magnification.
Colony formation assay
In brief, cells were plated into 6-well plates at a density of 2,000 cells per well and maintained in an incubator at 37 ℃ and 5% CO2. The cells were allowed to grow for 7 days to allow colony formation with or without mitotane or monastrol (30 µM). When the colonies had grown to an appropriate size, 1 mL of 4% paraformaldehyde was used to fix the cells for 20 min, and crystal violet was used for staining for 10 min, followed by washing with phosphate buffered saline, drying to obtain the images, and then counting the number of colonies.
Statistical analysis
All data were expressed as means ± standard deviations. Data from multiple groups were analyzed using analysis of variance (ANOVA) followed by Student’s t-test or Wilcoxon rank-sum test for comparisons between two groups. Correlations between clinical characteristics and KIF11 expression were analyzed using logistic regression. Univariate and multivariate Cox regression analyses were performed to determine the relationship between the KIF11 levels and clinical parameters. Receiver operating characteristic (ROC) curves for survival were plotted using the Kaplan-Meier survival ROC package. Nomograms and calibration plots were constructed with the “rms” package. Survival analyses and c-index calculations were performed using the Kaplan-Meier “survival” package. The above analyses were performed using R software (3.6.3) and GraphPad Prism (6.0). Differences were considered statistically significant at P<0.05.
Results
KIF11 expression was significantly elevated in ACC tissues
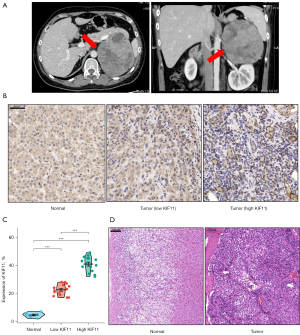
To explore the relationship between KIF11 expression and cancer progression, pan-cancer analyses was applied to compare the KIF11 expression levels in multiple cancer samples from the GTEx combined with TCGA and corresponding normal samples from the TCGA database. KIF11 was found to be highly expressed in most malignant tumors, including ACC, bladder urothelial carcinoma, breast infiltrating carcinoma, cervical squamous cell carcinoma, and adenocarcinoma (Figure 1A). The expression of KIF11 was then compared between 128 normal samples and 79 ACC samples from the TCGA_GTEx dataset. The KIF11 expression levels in the ACC tissues were found to be significantly higher than those in the normal adrenal tissues (P<0.001) (Figure 1B). Clinical information was collected from 30 patients with ACC who underwent surgery at Xiangya Hospital between January 2009 and January 2016. A typical enhanced computed tomography image of an ACC sample is shown in Figure 2A. Normal adrenal cortex tissues are neatly arranged into zona glomerulosa, zona fasciculata, and zona reticularis. In contrast, in the ACC tissues, the nucleus is large and deeply stained, the arrangement is disordered, and heterogeneity is evident. Furthermore, IHC staining to detect KIF11 expression showed that it was significantly higher in ACC samples than in normal adrenal tissues (Figure 2B,2C). The staining intensity was divided into ‘KIF11-high’ (greater than or equal to 30% positive nuclei) and ‘KIF11 low’ (less than 30% positive nuclei) groups, for comparison with clinical severity (given below). H&E staining of normal adrenal tissues and an ACC sample is shown in Figure 2D. The resulting data indicate that KIF11 expression is significantly elevated in ACC tissues, and that KIF11 may be an important marker of ACC.

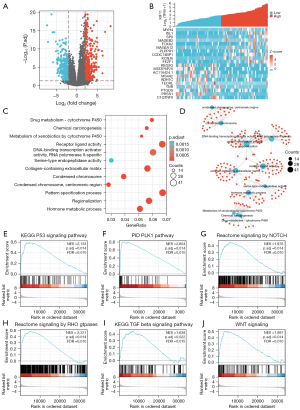
Identification of genes corelated with KIF11 in ACC
The ACC samples from the TCGA database were divided into two groups according to their KIF11 expression, and the 40 samples from the KIF11-high group were compared with 39 samples from the KIF11-low group used as a control. A total of 941 DEGs (395 downregulated and 546 upregulated) showed statistically significant differences between the two cohorts (adjusted P<0.05, |Log2 fold change| >2.0) (Figure 3A). The relative expression levels of the top 20 DEGs between the two cohorts are shown in Figure 3B, including 10 up-regulated and 10 down-regulated genes. Metascape was then used for GO and KEGG analyses of the DEGs. The top 12 GO enrichment terms were identified (Figure 3C), which included receptor-ligand activity, pattern specification process, DNA-binding transcription activator activity, RNA polymerase II-specific, and collagen-containing extracellular matrix. The correlations among the top 12 enriched terms from the GO analysis are shown as a network in Figure 3D. GSEA was conducted to identify the KIF11-related signaling pathways in ACC using the TCGA data. Several pathways and biological processes were found to be differentially enriched in the KIF11-high ACC group, including the activated KEGG-p53, PID-PLK1, Notch, Rho-GTPase, KEGG-TGFβ, and Wnt signaling pathways (Figure 3E-3J).
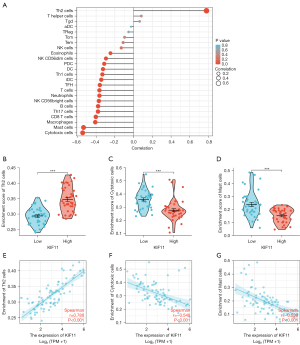
The correlation between KIF11 expression and immune infiltration
The correlation between the expression level of KIF11 and immune cell infiltration level quantified by ssGSEA was analyzed by Spearman correlation. The expression of KIF11 was positively correlated with the abundance of Th2 cells, and negatively correlated with the abundance of Cytotoxic cells, Mast cells, and Macrophages etc. (Figure 4).
Increased KIF11 was associated with shorter survival time and clinicopathological variables in patients with ACC
ROC analysis was used to analyze the ability of KIF11 to discriminate between ACC and normal adrenal tissues. The area under the curve (AUC) of KIF11 was 0.866, suggesting that KIF11 may be a potential diagnostic marker for ACC (Figure 5A). Patients in the KIF11-high group had poorer OS [hazard ratio (HR) =12.61, P<0.001], DSS (HR =12.08, P<0.001), and PFI (HR =5.96, P<0.001) as compared to those in the KIF11-low group (Figure 5B-5D). The relationship between KIF11 expression and the clinicopathological variables was then analyzed. Increased KIF11 demonstrated a significantly positive correlation with the T stages (T4 vs. T1, P<0.01; T4 vs. T2, P<0.001), M stages (M1 vs. M0, P<0.001), pathological stages (Stage IV vs. Stage I, P<0.001; Stage IV vs. Stage II, P<0.001; Stage IV vs. Stage III, P<0.05), primary therapy outcome [progressive disease (PD) and stable disease (SD) and partial response (PR) vs. complete response (CR), P<0.001], residual tumor (R1 and R2 vs. R0, P<0.001), venous invasion (present vs. absent, P<0.01), and invasion of the tumor capsule (present vs. absent, P<0.05) (Figure 5E-5L and Table 1).
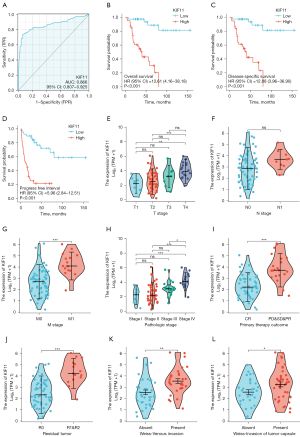
Table 1
| Characteristic | Low expression of KIF11 (n=39) | High expression of KIF11 (n=40) | P |
|---|---|---|---|
| T stage, n (%) | 0.002 | ||
| T1 | 7 (9.1) | 2 (2.6) | |
| T2 | 26 (33.8) | 16 (20.8) | |
| T3 | 3 (3.9) | 5 (6.5) | |
| T4 | 3 (3.9) | 15 (19.5) | |
| N stage, n (%) | 0.014 | ||
| N0 | 38 (49.4) | 30 (39.0) | |
| N1 | 1 (1.3) | 8 (10.4) | |
| M stage, n (%) | <0.001 | ||
| M0 | 38 (49.4) | 24 (31.2) | |
| M1 | 1 (1.3) | 14 (18.2) | |
| Pathologic stage, n (%) | <0.001 | ||
| Stage I | 7 (9.1) | 2 (2.6) | |
| Stage II | 25 (32.5) | 12 (15.6) | |
| Stage III | 6 (7.8) | 10 (13.0) | |
| Stage IV | 1 (1.3) | 14 (18.2) | |
| Tumor status, n (%) | <0.001 | ||
| Tumor free | 31 (40.3) | 8 (10.4) | |
| With tumor | 8 (10.4) | 30 (39.0) | |
| Primary therapy outcome, n (%) | <0.001 | ||
| PD | 1 (1.5) | 17 (25.4) | |
| SD | 1 (1.5) | 1 (1.5) | |
| PR | 1 (1.5) | 0 (0.0) | |
| CR | 34 (50.7) | 12 (17.9) | |
| Gender, n (%) | 0.928 | ||
| Female | 23 (29.1) | 25 (31.6) | |
| Male | 16 (20.3) | 15 (19.5) | |
| Age, n (%) | 0.571 | ||
| ≤50 years | 22 (27.8) | 19 (24.1) | |
| >50 years | 17 (21.5) | 21 (26.6) | |
| Age (years), median (IQR) | 48 (33.5, 56.5) | 52 (35.5, 61.0) | 0.662 |
| Residual tumor, n (%) | <0.001 | ||
| R0 | 34 (48.6) | 21 (30.0) | |
| R1 | 0 (0.0) | 6 (8.6) | |
| R2 | 1 (1.4) | 8 (11.4) | |
| Laterality, n (%) | 0.897 | ||
| Left | 23 (29.1) | 22 (27.8) | |
| Right | 16 (20.3) | 18 (22.8) | |
KIF11, kinesin family member 11; TCGA, The Cancer Genome Atlas; PD, progressive disease; SD, stable disease; PR, partial response; CR, complete response.
Univariate logistic regression analysis showed that KIF11 expression was a categorical dependent variable associated with poor prognostic clinicopathological characteristics (Table 2). High expression of KIF11 in the ACC was positively associated with the T stages [odds ratio (OR) = 6.111 for T3 and T4 vs. T1 and T2], N stages (OR =10.133 for T3 and T4 vs. T1 and T2), M stages (OR =22.167 for T3 and T4 vs. T1 and T2), pathological stages (OR =7.837 for Stage III and IV vs. Stage I and II), residual tumor (OR =22.667 for R1 and R2 vs. R0), tumor status (OR =14.531 for tumor vs. tumor free), primary therapy outcome (OR =17.000 for PD and SD and PR vs. CR), venous invasion (OR =4.889 for present vs. absent), and invasion of the tumor capsule (OR =2.674 present vs. absent). These results suggest that ACC in the KIF11-high group is prone to progression.
Table 2
| Characteristics | Total (N) | OR | P |
|---|---|---|---|
| T stage (T3 & T4 vs. T1 & T2) | 77 | 6.111 (2.179–19.308) | <0.001 |
| N stage (N1 vs. N0) | 77 | 10.133 (1.723–193.353) | 0.033 |
| M stage (M1 vs. M0) | 77 | 22.167 (4.059–414.648) | 0.004 |
| Pathologic stage (Stage III & Stage IV vs. Stage I & Stage II) | 77 | 7.837 (2.861–23.806) | <0.001 |
| Residual tumor (R1 & R2 vs. R0) | 70 | 22.667 (4.102–425.787) | 0.004 |
| Tumor status (with tumor vs. tumor free) | 77 | 14.531 (5.088–46.742) | <0.001 |
| Primary therapy outcome (PD & SD & PR vs. CR) | 67 | 17.000 (4.762–82.393) | <0.001 |
| Weiss-venous invasion (present vs. absent) | 70 | 4.889 (1.810–14.161) | 0.002 |
| Weiss-invasion of tumor capsule (present vs. absent) | 73 | 2.674 (1.040–7.164) | 0.045 |
KIF11, kinesin family member 11; OR, odds ratio; PD, progressive disease; SD, stable disease; PR, partial response; CR, complete response.
In addition, we analyzed the clinical data of 30 patients with ACC who underwent surgery at Xiangya Hospital between January 2009 and January 2016 (Figure 6, Table 3). The patients were divided into two groups (KIF11-high and KIF11-low) according to the expression of KIF11 detected by IHC. The results showed that the overexpression of KIF11 had a significantly positive correlation with tumor size (7.22±0.35 vs. 8.35±0.36, P=0.047), T stages (T3 & T4 vs. T1 & T2, P=0.021), pathological stages (Stage III & IV vs. Stage I & II, P=0.009), and tumor recurrence rate within 2 years (P=0.025). Patients in the KIF11-high group had poorer OS (Figure 6, HR =3.58, P=0.03).
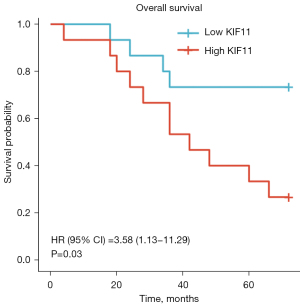
Table 3
| Characteristic | Low expression of KIF11 (n=15) | High expression of KIF11 (n=15) | P |
|---|---|---|---|
| Gender, n (%) | 0.715 | ||
| Female | 7 (23.3) | 9 (30.0) | |
| Male | 8 (26.7) | 6 (20.0) | |
| Age, n (%) | 1.000 | ||
| ≤50 years | 5 (16.7) | 4 (13.3) | |
| >50 years | 10 (33.3) | 11 (36.7) | |
| Laterality, n (%) | 0.272 | ||
| Left | 6 (20.0) | 10 (33.3) | |
| Right | 9 (30.0) | 5 (16.7) | |
| Tumor size (cm), mean ± SD | 7.2±0.4 | 8.4±0.4 | 0.037 |
| T stage, n (%) | 0.021 | ||
| T1 & T2 | 13 (43.3) | 6 (20.0) | |
| T3 & T4 | 2 (6.7) | 9 (30.0) | |
| N stage, n (%) | 0.390 | ||
| N0 | 13 (43.3) | 10 (33.3) | |
| N1 | 2 (6.7) | 5 (16.7) | |
| M stage, n (%) | 0.598 | ||
| M0 | 14 (46.7) | 12 (43.3) | |
| M1 | 1 (3.3) | 3 (6.7) | |
| Pathologic stage, n (%) | 0.009 | ||
| Stage I & Stage II | 12 (40.0) | 4 (13.3) | |
| Stage III & Stage IV | 3 (10.0) | 11 (36.7) | |
| Tumor recurrence (within 2 years), n | 3 | 10 | 0.025 |
KIF11, kinesin family member 11; SD, standard deviation.
Blocking KIF11 inhibits NCI-H295R cell proliferation and invasion
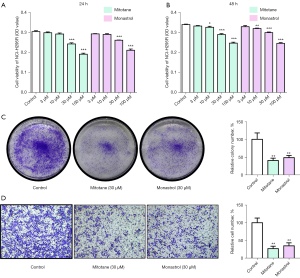
We found that many compounds tested against the protein target KIF11 by using PubChem (Table S1). To prove that KIF11 is a druggable target, we examined the effect of the KIF11 one of the specific inhibitor monastrol on the proliferation and invasion ability of NCI-H295R cells. Mitotane was also used, as it is the standard treatment used for ACC to kill tumor cells. The CCK-8 assay showed that NCI-H295R cell treated with mitotane and monastrol (30 µM/100 µM) for 24 hours and 48 hours exhibited obvious decreases in their proliferation (Figure 7A,7B). Consequently, 30 µM of the mitotane and monastrol was then used for further experiments. The colony formation assay also showed decreased proliferation of NCI-H295R cell after treatment with 30 µM of mitotane and monastrol (Figure 7C). The Transwell invasion assay revealed that the mitotane and monastrol treatments significantly decreased NCI-H295R cell invasion when compared to the control group (Figure 7D). Overall, the results demonstrated that monastrol could inhibit NCI-H295R cell proliferation and invasion in vitro, similar to mitotane, which is the mainstay of adjuvant treatment for ACC.
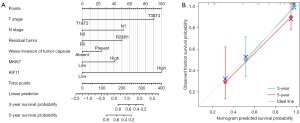
Nomogram for predicting OS in ACC patients based on KIF11 expression
In view of the prognostic value of KIF11 in ACC, we created a nomogram for predicting the 3- and 5-year survival (Figure 8). The tumor status, invasion of the tumor capsule, and the expression of Ki67 have been reported to be associated with the incidence or prognosis of ACC (5,23,24). Therefore, these parameters were included in the predictive model (Figure 8A). We also analyzed the prediction efficiency of the nomogram, and the results indicated that the C-index of the model was 0.891, which suggests that its prediction efficiency was moderately accurate.
Discussion
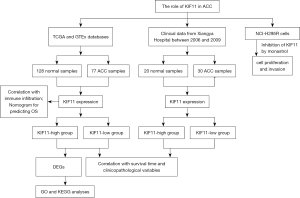
To the best of our knowledge, this is the first study focusing on the potential effects of KIF11 in relation to ACC. Bioinformatics analysis using the TCGA and GTEx databases and 7 years of statistics from Xiangya Hospital demonstrated that KIF11 may be a potential prognostic biomarker for ACC. Increased KIF11 expression in ACC was found to be associated with clinical pathological characteristics, shorter survival time, and poor prognosis. Monastrol was also assessed and the results showed that KIF11 is a druggable target for limiting ACC cell proliferation and invasion. The flow chart of this study is showed in Figure 9.
Despite a better understanding of the pathophysiology of ACC and the gradual appearance of more treatment options, the prognosis of ACC remains poor. Etoposide, doxorubicin, cisplatin, and mitotane (EDP-M) have been established as first-line therapies for metastatic ACC. However, this treatment regimen shows significant toxicity potential in patients (6). In recent years, drugs targeting the IGF pathway, mTOR or tyrosine kinase inhibitors, and immunotherapy-targets have been extensively studied in relation to ACC (8,25). However, most of these drugs are found to have relatively low response rates and results in no significant improvements to OS rates for ACC (6,26,27).
Cell proliferation is one of the most important hallmarks of cancer development and progression. Correct alignment of the mitotic spindle during cell division is crucial for the determination of cell fate, tissue organization, and development (28). The accurate segregation of chromosomes is mediated by microtubule-based mitotic spindles and approximately 200 essential microtubule-associated proteins (29). KIF11, which is perhaps the simplest player in the mitotic spindle assembly, is a plus-end-directed motor localized to interpolar spindle microtubules and spindle poles (10). KIF11 has been reported to play an essential role in centrosome separation by cross-linking the microtubules in the mitotic spindle (30). The suppression of KIF11 increases the proportion of cells in the G2/M phase and sub-G1 phase, indicating that it has a vital role in G2/M phase transition and cell cycle checkpoints (31). To investigate the functions of KIF11 in ACC, GO, GSEA, and single-sample GSEA analyses with TCGA data were performed. The results revealed that the KEGG-p53, PID-PLK1, Notch, Rho-GTPase, KEGG-TGFβ, and Wnt signaling pathways were differentially enriched in the KIF11-high group. Accumulating evidence has indicated that these signaling pathways regulate many aspects of cancer biology (32-37).
In recent years, KIF11 has attracted the attention of researchers as it has been shown to have a therapeutic role in a variety of tumors, including oral cancer (31), meningioma (18), glioblastoma (38), hepatocellular carcinoma (12), breast cancer (39), gallbladder cancer (40), and lung cancer (41). A KIF11 inhibitor was shown to demonstrate better efficacy in hematological malignancies due to the higher proliferative rates of blood cancers and off-target activities of anti-mitotic agents against oncogenic drivers (42,43). However, clinical research studies on KIF11 inhibitor found that mitotic inhibition alone does not appear to be sufficient to achieve significant antitumor effect (44,45). Furthermore, combination therapies may be more promising due to the multiple synergistic interactions between anti-mitotic and anticancer drugs (43,46). In this study, KIF11 inhibitor monastrol was applied to treat NCI-H295R cell and cell proliferation and invasion were significantly suppressed. However, further research is required to fully understand the role and mechanism of monastrol in relation to ACC.
While the results suggest that KIF11 may be a predictor of poor prognosis as well as a novel therapeutic target for ACC, some limitations exist in this study. Firstly, the overall sample size used for the RNA-seq analysis was small. The number of patients enrolled from Xiangya Hospital was also small due to the low incidence of ACC, and the follow-up time was short. Therefore, future research with a longer period of follow-up is worth investigated. Secondly, a prospective study should be performed in the future to avoid bias arising from the retrospective nature of the current study. Thirdly, the detailed mechanism of how KIF11 impacts cell proliferation and invasion in ACC should be elucidated. Finally, additional strategies to assess the role of KIF11 in ACC, including the use of KIF11 knockout cells and animal experiments, may be required in the future. These limitations need to be addressed in future studies.
Conclusions
In conclusion, this is the first study to report that high KIF11 expression levels are significantly associated with the progression of ACC and resulting in poor survival rates. Furthermore, KIF11 expression appears to be a novel and independent prognostic marker in ACC. KIF11 is a key protein involved in cell proliferation and invasion, and may thus serve as a potential novel therapeutic target for ACC.
Acknowledgments
We would like to thank Editage (www.editage.cn) for English language editing.
Funding: This work was supported by grants from Xiangya Clinical Data System of Central South University (to Xiang Chen).
Footnote
Reporting Checklist: The authors have completed the STROBE reporting checklist. Available at https://tau.amegroups.com/article/view/10.21037/tau-22-706/rc
Data Sharing Statement: Available at https://tau.amegroups.com/article/view/10.21037/tau-22-706/dss
Peer Review File: Available at https://tau.amegroups.com/article/view/10.21037/tau-22-706/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tau.amegroups.com/article/view/10.21037/tau-22-706/coif). XC reports the funding from Xiangya Clinical Data System of Central South University. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of the Xiangya Hospital, China (No. 202109889) and all samples and clinical information used in this study were obtained with informed consent from patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Torti JF, Correa R. Adrenal Cancer. Treasure Island (FL): StatPearls, 2022.
- Hasebe M, Shibue K, Honjo S, et al. Adrenocortical carcinoma. QJM 2022;115:43-4. [Crossref] [PubMed]
- Stojadinovic A, Ghossein RA, Hoos A, et al. Adrenocortical carcinoma: clinical, morphologic, and molecular characterization. J Clin Oncol 2002;20:941-50. [Crossref] [PubMed]
- Fassnacht M, Allolio B. Clinical management of adrenocortical carcinoma. Best Pract Res Clin Endocrinol Metab 2009;23:273-89. [Crossref] [PubMed]
- Fassnacht M, Johanssen S, Quinkler M, et al. Limited prognostic value of the 2004 International Union Against Cancer staging classification for adrenocortical carcinoma: proposal for a Revised TNM Classification. Cancer 2009;115:243-50. [Crossref] [PubMed]
- Subramanian C, McCallister R, Kuszynski D, et al. Re-Evaluation of Combinational Efficacy and Synergy of the Italian Protocol In Vitro: Are We Truly Optimizing Benefit or Permitting Unwanted Toxicity? Biomedicines 2021;9:1190. [Crossref] [PubMed]
- Pittaway JFH, Guasti L. Pathobiology and genetics of adrenocortical carcinoma. J Mol Endocrinol 2019;62:R105-19. [Crossref] [PubMed]
- Fassnacht M, Dekkers OM, Else T, et al. European Society of Endocrinology Clinical Practice Guidelines on the management of adrenocortical carcinoma in adults, in collaboration with the European Network for the Study of Adrenal Tumors. Eur J Endocrinol 2018;179:G1-G46. [Crossref] [PubMed]
- Shi J, Mitchison TJ. Cell death response to anti-mitotic drug treatment in cell culture, mouse tumor model and the clinic. Endocr Relat Cancer 2017;24:T83-96. [Crossref] [PubMed]
- Garcia-Saez I, Skoufias DA. Eg5 targeting agents: From new anti-mitotic based inhibitor discovery to cancer therapy and resistance. Biochem Pharmacol 2021;184:114364. [Crossref] [PubMed]
- Zhou J, Chen WR, Yang LC, et al. KIF11 Functions as an Oncogene and Is Associated with Poor Outcomes from Breast Cancer. Cancer Res Treat 2019;51:1207-21. [Crossref] [PubMed]
- Hu ZD, Jiang Y, Sun HM, et al. KIF11 Promotes Proliferation of Hepatocellular Carcinoma among Patients with Liver Cancers. Biomed Res Int 2021;2021:2676745. [Crossref] [PubMed]
- Wang H, Li S, Liu B, et al. KIF11: A potential prognostic biomarker for predicting bone metastasis-free survival of prostate cancer. Oncol Lett 2022;24:312. [Crossref] [PubMed]
- Mo XC, Zhang ZT, Song MJ, et al. Screening and identification of hub genes in bladder cancer by bioinformatics analysis and KIF11 is a potential prognostic biomarker. Oncol Lett 2021;21:205. [Crossref] [PubMed]
- Jin Q, Dai Y, Wang Y, et al. High kinesin family member 11 expression predicts poor prognosis in patients with clear cell renal cell carcinoma. J Clin Pathol 2019;72:354-62. [Crossref] [PubMed]
- Imai T, Oue N, Nishioka M, et al. Overexpression of KIF11 in Gastric Cancer with Intestinal Mucin Phenotype. Pathobiology 2017;84:16-24. [Crossref] [PubMed]
- Schneider MA, Christopoulos P, Muley T, et al. AURKA, DLGAP5, TPX2, KIF11 and CKAP5: Five specific mitosis-associated genes correlate with poor prognosis for non-small cell lung cancer patients. Int J Oncol 2017;50:365-72. [Crossref] [PubMed]
- Jungwirth G, Yu T, Moustafa M, et al. Identification of KIF11 As a Novel Target in Meningioma. Cancers (Basel) 2019;11:545. [Crossref] [PubMed]
- Vivian J, Rao AA, Nothaft FA, et al. Toil enables reproducible, open source, big biomedical data analyses. Nat Biotechnol 2017;35:314-6. [Crossref] [PubMed]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [Crossref] [PubMed]
- Yu G, Wang LG, Han Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 2012;16:284-7. [Crossref] [PubMed]
- Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545-50. [Crossref] [PubMed]
- Kim Y, Margonis GA, Prescott JD, et al. Nomograms to Predict Recurrence-Free and Overall Survival After Curative Resection of Adrenocortical Carcinoma. JAMA Surg 2016;151:365-73. [Crossref] [PubMed]
- Zlatibor L, Paunovic I, Zivaljevic V, et al. Prognostic significance of immunohistochemical markers in adrenocortical carcinoma. Acta Chir Belg 2020;120:23-9. [Crossref] [PubMed]
- Altieri B, Ronchi CL, Kroiss M, et al. Next-generation therapies for adrenocortical carcinoma. Best Pract Res Clin Endocrinol Metab 2020;34:101434. [Crossref] [PubMed]
- Pereira SS, Monteiro MP, Costa MM, et al. IGF2 role in adrenocortical carcinoma biology. Endocrine 2019;66:326-37. [Crossref] [PubMed]
- Baudin E, Pellegriti G, Bonnay M, et al. Impact of monitoring plasma 1,1-dichlorodiphenildichloroethane (o,p'DDD) levels on the treatment of patients with adrenocortical carcinoma. Cancer 2001;92:1385-92. [Crossref] [PubMed]
- Noatynska A, Gotta M, Meraldi P. Mitotic spindle (DIS)orientation and DISease: cause or consequence? J Cell Biol 2012;199:1025-35. [Crossref] [PubMed]
- Petry S. Mechanisms of Mitotic Spindle Assembly. Annu Rev Biochem 2016;85:659-83. [Crossref] [PubMed]
- Wojcik EJ, Buckley RS, Richard J, et al. Kinesin-5: cross-bridging mechanism to targeted clinical therapy. Gene 2013;531:133-49. [Crossref] [PubMed]
- Daigo K, Takano A, Thang PM, et al. Characterization of KIF11 as a novel prognostic biomarker and therapeutic target for oral cancer. Int J Oncol 2018;52:155-65. [PubMed]
- Meurette O, Mehlen P. Notch Signaling in the Tumor Microenvironment. Cancer Cell 2018;34:536-48. [Crossref] [PubMed]
- Taciak B, Pruszynska I, Kiraga L, et al. Wnt signaling pathway in development and cancer. J Physiol Pharmacol 2018; [PubMed]
- Zhao H, Wei J, Sun J. Roles of TGF-β signaling pathway in tumor microenvirionment and cancer therapy. Int Immunopharmacol 2020;89:107101. [Crossref] [PubMed]
- Bykov VJN, Eriksson SE, Bianchi J, et al. Targeting mutant p53 for efficient cancer therapy. Nat Rev Cancer 2018;18:89-102. [Crossref] [PubMed]
- Liu Z, Sun Q, Wang X. PLK1, A Potential Target for Cancer Therapy. Transl Oncol 2017;10:22-32. [Crossref] [PubMed]
- Zeng RJ, Zheng CW, Chen WX, et al. Rho GTPases in cancer radiotherapy and metastasis. Cancer Metastasis Rev 2020;39:1245-62. [Crossref] [PubMed]
- Venere M, Horbinski C, Crish JF, et al. The mitotic kinesin KIF11 is a driver of invasion, proliferation, and self-renewal in glioblastoma. Sci Transl Med 2015;7:304ra143. [Crossref] [PubMed]
- Wang B, Yu J, Sun Z, et al. Kinesin family member 11 is a potential therapeutic target and is suppressed by microRNA-30a in breast cancer. Mol Carcinog 2020;59:908-22. [Crossref] [PubMed]
- Wei D, Rui B, Qingquan F, et al. KIF11 promotes cell proliferation via ERBB2/PI3K/AKT signaling pathway in gallbladder cancer. Int J Biol Sci 2021;17:514-26. [Crossref] [PubMed]
- Kato T, Lee D, Huang H, et al. Personalized siRNA-Nanoparticle Systemic Therapy using Metastatic Lymph Node Specimens Obtained with EBUS-TBNA in Lung Cancer. Mol Cancer Res 2018;16:47-57. [Crossref] [PubMed]
- Borisa AC, Bhatt HG. A comprehensive review on Aurora kinase: Small molecule inhibitors and clinical trial studies. Eur J Med Chem 2017;140:1-19. [Crossref] [PubMed]
- Tischer J, Gergely F. Anti-mitotic therapies in cancer. J Cell Biol 2019;218:10-1. [Crossref] [PubMed]
- Kantarjian HM, Padmanabhan S, Stock W, et al. Phase I/II multicenter study to assess the safety, tolerability, pharmacokinetics and pharmacodynamics of AZD4877 in patients with refractory acute myeloid leukemia. Invest New Drugs 2012;30:1107-15. [Crossref] [PubMed]
- Infante JR, Patnaik A, Verschraegen CF, et al. Two Phase 1 dose-escalation studies exploring multiple regimens of litronesib (LY2523355), an Eg5 inhibitor, in patients with advanced cancer. Cancer Chemother Pharmacol 2017;79:315-26. [Crossref] [PubMed]
- Gutteridge RE, Ndiaye MA, Liu X, et al. Plk1 Inhibitors in Cancer Therapy: From Laboratory to Clinics. Mol Cancer Ther 2016;15:1427-35. [Crossref] [PubMed]


