
Comparisons of the diagnostic accuracy across prostate health index, prostate health index density, and percentage free prostate-specific antigen for clinically significant prostate cancer: a prospective diagnostic study
Highlight box
Key findings
• We found prostate health index density was more accurate in predicting positive biopsies with clinically significant prostate cancer in patients who have a prostate-specific antigen (PSA) level of 4–10 ng/mL.
What is known and what is new?
• Early diagnosis of clinically significant prostate cancer (csPCa) is challenging, and biopsy decision determined by conventional PSA is inaccurate and leads to excessive unnecessary biopsies;
• Prostate health index (PHI) is more accurate in screening positive biopsies. However, studies on PHI density (PHID) for predicting positive biopsy with csPCa have produced mixed results.
What is the implication, and what should change now?
• PHI should be used as routine protocol instead of conventional PSA in screening potential positive biopsies with csPCa to avoid unnecessary biopsies. PHID potentially outperforms PHI in patients with PSA in the gray zone in this setting.
Introduction
The serum prostate-specific antigen (PSA) and its derivative, percentage free PSA (%fPSA), have been widely used as biochemical markers for prostate cancer (PCa) screening in the early stages (1,2). However, its application has been much debated due to its poor specificity, especially in patients with PSA in the gray zone (PSA 4–10 ng/mL) (3). In a multicenter study, a total of 1,362 patients from 4 different study sites who had total PSA (tPSA) values of 1.6–8.0 µg/L were enrolled to evaluate the diagnostic performance of tPSA and %fPSA for detecting prostate cancer (4). The results showed that the diagnostic accurancy of tPSA and %fPSA were not satisfactory [area under the curve (AUC): 0.56 and 0.61, respectively]. Moreover, the result of another prospective, multicenter study indicated that the positive predictive value of %fPSA was only 0.325 for men with high-grade PCa (Gleason Score ≥7) (5). As a result, a large number of patients have undergone unnecessary prostate biopsies or been diagnosed with a nonclinically significant PCa which would not have affected the patient during the natural course of his lifetime. Furthermore, about 2% of patients have post-biopsy complications, such as infection, bleeding, or voiding difficulty (6). Such overdiagnosis and overtreatment not only deplete medical resources, but also harm the patients. Therefore, it is urgent to develop a novel biomarker to help the patients to avoid unnecessary prostate biopsies.
The isoform [-2]proPSA (p2PSA), which is a truncated variant of proPSA, is a relatively new serum marker for the early diagnosis of PCa (7). P2PSA derivatives, percentage of p2PSA (%p2PSA) and prostate health index (PHI), are calculated based on p2PSA and are superior to free PSA (fPSA) in diagnosing and predicting PCa (8,9). PHI was approved by the Chinese Food and Drug Administration, and its performance compared to conventional markers has not been fully studied in the Chinese population, which was the primary goal of this study. In addition, current guidelines recommend the use of PSA density (PSAD) to further improve the accuracy of PSA screening (10), and thus we hypothesized that PHI density (PHID), like PSAD, could also outperform PHI. However, the conclusions from published studies remain controversial (11-13). In a study including a large Caucasian group with 1,446 men from a single-center, PHID showed only a small advantage in comparison with PHI alone. And in smaller prostates, PHI even outperformed PHID (11). Moreover, Friedl et al. assessed the diagnostic performance of PHI and PHID in 112 males (12). And the results indicated that the AUC value of PHI (0.79) was higher than PHID (0.77). Similarly, in a prospective, observational multicenter study of two prostate biopsy cohorts from Asia, PHID did not improve the predictive ability of PHI for either PCa or clinically significant prostate cancer (csPCa) (13). Therefore, these controversial results promote us to investigate the diagnostic performance of PHID compared to PHI and %fPSA in predicting csPCa, especially in patients with PSA in the gray zone. We present the following article in accordance with the STARD reporting checklist (available at https://tau.amegroups.com/article/view/10.21037/tau-23-80/rc).
Methods
General information
This study was a prospective, observational single-center study in a prostate biopsy cohort at the First Hospital of Shanxi Medical University between September 2019 and December 2020. Serum samples and clinicopathological features were prospectively obtained from each patient who underwent prostate biopsy. The inclusion criteria were patients with indications for prostate biopsy [PSA >4 ng/mL, prostate magnetic resonance imaging (MRI) or ultrasound clearly suggesting an occupying lesion, etc.]. The exclusion criteria were: (I) patients with a history of other malignancies, (II) patients with interfering factors affecting PSA levels (related medical operations, use of 5α-reductase inhibitors, and history of acute urinary tract infection within 3 months), and (III) patients with missing diagnostic data. Out of 434 specimens, a total of 296 patients were finally enrolled in this study. Further subgroup analysis was performed on patients whose rectal examinations were negative and PSA values were between 4 and 10 ng/mL. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The protocol for this study was approved by the Ethics Committee of the First Hospital of Shanxi Medical University (approval No. K040). And all the participants gave informed consent before taking part in this study.
Research methods
Specimen collection and testing
Venous blood samples were collected immediately before transperineal prostate biopsy, processed within 3 hours, and stored at −20 to −80 ℃ until further analyses. Blood samples were processed using the Access 2 immunoassay system (DxI 800; Beckman Coulter, Brea, CA, USA). tPSA, fPSA, and p2PSA were measured according to Hybritech standards. PHI was calculated according to the formula PHI = p2PSA/fPSA × √tPSA, and PHID was calculated according to the formula PHID = PHI/PV. Preoperative prostate volume (PV) was measured by multiparametric MRI (mpMRI) of the prostate.
Prostate biopsy and pathology
All patients underwent mpMRI cognitive fusion plus rectal ultrasound-guided transperineal prostate biopsy. Depending on the PV and the location of the lesions suggested by mpMRI [prostate imaging-reporting and data systems (PI-RADs) score >3], 12–14 needles of systematic biopsy and targeted biopsy were performed (1–2 needles for each area with PI-RADs score >3). Transperineal prostate biopsy specimens were analyzed by an experienced urologic pathologist who did not know the results of the blood sample testing. PCa was given a grade according to the Gleason score: a score of ≥7 indicated csPCa, and a score of <7 indicated non-csPCa or no PCa findings.
Statistical methods
All data were processed and analyzed using SPSS 26.0 statistical software. Quantitative data conforming to normal distribution are presented as mean ± standard deviation, and the differences between groups were compared using independent samples t-test. Quantitative data that were not normally distributed are presented as median and interquartile range (IQR), and the differences between groups were compared using the Mann-Whitney test. The receiver operating characteristic (ROC) curve was plotted, and the AUC value was measured to evaluate the diagnostic performance. Statistical differences between AUCs were evaluated using the DeLong method (14). The 25th percentile value of PHID was considered as the cut-off value to calculate the sensitivity and specificity. Furthermore, we also investigated the prevalence of csPCa between 25th and 75th percentile value of PHID and >75th percentile value of PHID. A two-tailed P<0.05 was defined as statistically significant.
Results
General information about the study population
Based on the inclusion and exclusion criteria, a total of 296 patients were finally enrolled in this study. The clinicopathological characteristics of the patients are presented in Table 1. Among the 76 patients (25.68%) with PCa, 54 patients with Gleason score ≥7 were diagnosed with csPCa. We classified a total of 242 patients, including 22 patients with Gleason score <7 and 220 patients (74.32%) without malignancy, as the non-csPCa group. The differences between the csPCa and the non-csPCa groups were statistically significant with respect to patient age, tPSA, p2PSA, PHI, and PHID (Table 1). Furthermore, in the subgroup analysis, a total of 198 patients exhibited PSA in the gray zone.
Table 1
| Feature | Overall (n=296) | Non-csPCa (n=242) (BPH and Gleason <7) | csPCa (n=54) (Gleason ≥7) | P |
|---|---|---|---|---|
| Age (years), mean ± SD | 67.5±7.9 | 66.8±7.8 | 70.4±8.0 | 0.030* |
| Prostate volume (mL), median (IQR) | 44.76 (35.84–57.20) | 44.96 (35.96–59.00) | 43.92 (32.76–51.98) | 0.357 |
| tPSA (ng/mL), median (IQR) | 7.94 (5.90–11.88) | 7.67 (5.63–10.27) | 11.00 (7.21–18.72) | 0.001* |
| %fPSA (%), median (IQR) | 16.89 (12.03–22.25) | 18.21 (12.18–22.87) | 13.66 (11.78–17.56) | 0.099 |
| p2PSA (ng/L), median (IQR) | 15.70 (10.03–25.85) | 13.74 (9.02–22.05) | 30.21 (17.53–95.66) | <0.001* |
| PHI, median (IQR) | 32.52 (23.81–52.75) | 29.41 (21.30–42.78) | 64.14 (40.52–136.23) | <0.001* |
| PHID, median (IQR) | 0.65 (0.51–1.06) | 0.59 (0.47–0.83) | 1.62 (1.02–3.04) | <0.001* |
*, P<0.05 was significant. csPCa, clinically significant prostate cancer; BPH, benign prostatic hyperplasia; IQR, interquartile range; tPSA, total prostate-specific antigen; %fPSA, percentage free prostate-specific antigen; p2PSA, isoform [-2]proPSA; PHI, prostate health index; PHID, prostate health index density; SD, standard deviation.
Diagnostic performance of PHI and PHID for csPCa in all patients
The results of the diagnostic analysis showed that, the AUC value for PHI (0.867) was higher than that for p2PSA (0.805) and the other parameters, including the value for tPSA (0.699) and %fPSA (0.602). PHID had a slightly higher (although not significantly different) AUC value than PHI, indicating that PHID had an ability to diagnose csPCa comparable to PHI (both P<0.05, see Table 2 and Figure 1).
Table 2
| Parameters | AUC | 95% CI | P |
|---|---|---|---|
| tPSA | 0.699 | 0.589–0.810 | 0.012* |
| %fPSA | 0.602 | 0.491–0.713 | <0.001* |
| p2PSA | 0.805 | 0.720–0.890 | 0.270 |
| PHI | 0.867 | 0.796–0.938 | – |
| PHID | 0.880 | 0.803–0.957 | 0.807 |
*, P<0.05 was significant. csPCa, clinically significant prostate cancer; AUC, area under the curve; CI, confidence interval; tPSA, total prostate-specific antigen; %fPSA, percentage free prostate-specific antigen; p2PSA, isoform [-2]proPSA; PHI, prostate health index; PHID, prostate health index density.
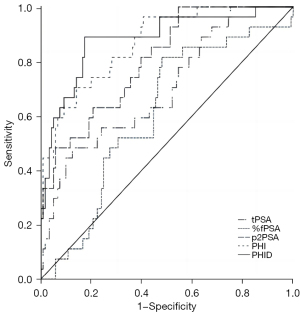
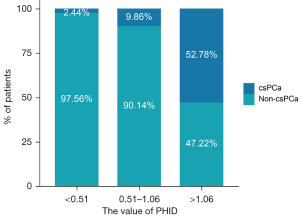
The median PHID value was 0.65 in all patients, and the 25th and 75th percentile values were 0.51 and 1.06. Fifty-two (96.30%) of the 54 csPCa patients who had a PHID value >0.51. The sensitivity of PHID for detecting csPCa was 96.30%, and the specificity was 33.06% with the cut-off value of 0.51. Using the 25th percentile of PHID as the cut-off value, a negative predictive value of 97.56% was achieved; the diagnosis was missed in only 2 (3.70%) patients with csPCa. As shown in Figure 2, the prevalence of csPCa increased significantly with increasing PHID values, with 2.44% of patients with PHID <0.51 developing csPCa, and the prevalence of csPCa increased to 9.86% in the PHID range of 0.51–1.06. In patients with PHID >1.06, 52.78% were diagnosed with csPCa.
Diagnostic performance of PHI and PHID in patients with PSA within 4–10 ng/mL
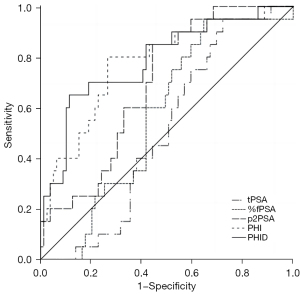
A biopsy decision within this specific PSA range is mostly difficult. Therefore, we additionally analyzed those 198 men with PSA values within 4–10 ng/mL, which included 40 cases in the csPCa group and 158 cases of PCa with Gleason <7 in the non-csPCa group. The patients were significantly older in the csPCa group compared to the non-csPCa group, with significantly higher levels of PSA, %fPSA, p2PSA, PHI, and PHID. Further comparison of the performance of each marker in predicting PCa in patients with PSA 4–10 ng/mL revealed the following: PHID (AUC: 0.788) > PHI (AUC: 0.777) > p2PSA (AUC: 0.691) (differences were significant, P=0.035, P=0.029). Compared to PHI, tPSA and %fPSA, with an AUC of 0.508 and 0.569, respectively, had poorer diagnostic power, and the difference was statistically significant (both P<0.05, see Table 3 and Figure 3). These results showed that PHID outperformed all the other PSA derivative markers in detecting csPCa in patients with PSA within the gray zone.
Table 3
| Parameters | AUC | 95% CI | P |
|---|---|---|---|
| tPSA | 0.508 | 0.391–0.625 | 0.001* |
| %fPSA | 0.569 | 0.449–0.689 | 0.014* |
| p2PSA | 0.691 | 0.579–0.802 | 0.029* |
| PHI | 0.777 | 0.662–0.892 | – |
| PHID | 0.788 | 0.672–0.904 | 0.035* |
*, P<0.05 was significant. PCa, prostate cancer; PSA, prostate-specific antigen; AUC, area under the curve; CI, confidence interval; tPSA, total prostate-specific antigen; %fPSA, percentage free prostate-specific antigen; p2PSA, isoform [-2]proPSA; PHI, prostate health index; PHID, prostate health index density.
Discussion
The treatment strategy for patients with nonsignificant PCa includes watchful waiting and active surveillance, with only csPCa patients requiring curative treatment (10). Therefore, the screening of csPCa becomes an important part of the treatment strategy (15). Studies have shown that approximately 75% of patients undergo unnecessary prostate biopsies based on PSA values only (16,17). Excessive biopsies lead to serious complications, such as urinary tract infections, pain, and hematuria (18). In addition, 1.3% of these patients experience complicated infections, and 3.9% even require further hospitalization, seriously affecting quality of life (19). The PSA is now widely used for screening PCa patients. And the application of %fPSA could improve the detection rate of PCa. However, studies have shown that the diagnostic efficacy of %fPSA for PCa is still limited at the time of initial biopsy. A meta-analysis by Huang et al. demonstrated that the sensitivity of %fPSA for predicting positive biopsy patients ranged from 0.5 to 0.94 [pooled sensitivity 0.70, 95% confidence interval (CI): 0.67–0.72], while the specificity ranged from 0.31 to 0.93 (pooled specificity 0.55, 95% CI: 0.57–0.60) (20). Our study results showed that PHI and PHID significantly outperformed %fPSA in predicting positive biopsy in patients with PSA in the gray zone and in predicting csPCa in all patients with an increased PSA.
In this study, we observed that 2.44% of patients with PHID <0.51 (< Q25) and 52.78% of patients with PHID >1.06 (> Q75) developed csPCa, which further demonstrated that as PHID increases, the detection rate of csPCa also increases. Subsequently, we further evaluated the predictive ability of PHI, PHID, and conventional indicators for csPCa. The sensitivity of PHID for detecting csPCa was 96.30%, and the specificity was 33.06% based on the 25th percentile (0.51) of PHID as the cutoff value, and only 2 (3.70%) patients with csPCa were missed. The AUC value of PHI and PHID for predicting csPCa were 0.867 and 0.880, respectively, which indicated the good diagnostic performance of PHI and PHID. In contrast, traditional tumor markers tPSA and %fPSA were both significantly inferior to PHI in predicting clinically significant PCa, with an AUC of 0.699 (P=0.012) and 0.602 (P<0.001), respectively. These outcomes were consistent with the findings of a study of 412 Taiwanese men conducted by Chiu et al. (21), whose results showed an AUC for tPSA, %fPSA, %p2PSA, PSAD, PHI, and PHID of 0.56, 0.63, 0.76, 0.74, 0.77, and 0.82, respectively, for csPCa detection in all patients with elevated PSA. Overall, the results of our study suggested that PHID and PHI had a high prediction rate for csPCa in all patients.
In terms of predicting PCa in patients with PSA in the gray zone, a European study in 2020 involving a large number of patients with PSA 1–8 ng/mL found that the diagnostic efficacy of PHID in predicting positive biopsy had an AUC of 0.819, which was superior to that of PHI, with an AUC of 0.789 (P=0.0219) (22). In this study, we found the same diagnostic performance in the gray zone of PSA 4–10 ng/mL, with the AUC of PHID higher than that for PHI in predicting biopsy with csPCa (AUC: 0.788 vs. 0.777, P=0.035). In contrast, conventional markers, including tPSA and %fPSA, had inferior diagnostic performance in patients with PSA in the gray zone (AUC: 0.508 and 0.569, respectively). In all, PHID had the best accuracy rate in predicting csPCa in patients whose PSA was within the gray zone. However, it is important to emphasize that, due to the small number of csPCa patients (n=40), the better performance of PHID compared wtih PHI in predicting csPCa in patients with PSA in the gray zone should be validated in the larger cohort. And it would be the goal and direction of our future research.
With their excellent performance, both PHI and PHID could serve as alternative markers for Gleason monitoring in PCa patients undergoing active surveillance as they can accurately predict the Gleason level of prostate biopsy results, thereby avoiding overdiagnosis and overtreatment. Furthermore, advances in diagnostic imaging technology have made it possible to identify lesions that are difficult to visualize using conventional methods (23). Therefore, it is expected that the combination of PHI and PHID and imaging investigation would allow patients to be free from undergoing painful prostate biopsies every year, leading to a significant improvement in their quality of life.
There were some limitations to this study. First, as a single-center study, the sample size was small, although all patients in this prospectively established patient pool decided to undergo prostate biopsy after preoperative analysis, enabling a more objective comparison of the diagnostic ability of PHI and PHID with traditional markers. Secondly, we used mpMRI cognitive fusion prostate biopsy instead of machine fusion, which might lead to some diagnostic limitations relating to technology. Therefore, further multicenter studies with large samples are needed to validate the results of this study.
Conclusions
Compared to %fPSA, PHI had higher accuracy in predicting csPCa in patients with elevated PSA. Compared to PHI, PHID resulted in comparable performance in these patients and did even better in patients with PSA in the gray zone and thus should form part of the treatment strategy for PCa patients.
Acknowledgments
Funding: This work was supported by the Scientific Research Funding Project of Returned Overseas Scholars of Shanxi Province (Grant No. 2021-160), the Basic Research Program of Shanxi Province (Grant No. 20210302123242), and the Beijing Bethune Charitable Foundation (Grant No. mnz1202029).
Footnote
Reporting Checklist: The authors have completed the STARD reporting checklist. Available at https://tau.amegroups.com/article/view/10.21037/tau-23-80/rc
Data Sharing Statement: Available at https://tau.amegroups.com/article/view/10.21037/tau-23-80/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tau.amegroups.com/article/view/10.21037/tau-23-80/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The protocol for this study was approved by the Ethics Committee of the First Hospital of Shanxi Medical University (approval No. K040). And all the participants gave informed consent before taking part in this study.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Narain TA, Sooriakumaran P. Beyond Prostate Specific Antigen: New Prostate Cancer Screening Options. World J Mens Health 2022;40:66-73. [Crossref] [PubMed]
- Oh JH, Chung HS, Kim MS, et al. Do prostate-specific antigen parameters have a similar role in predicting prostate cancer regardless of serum testosterone levels in men with gray-zone prostate-specific antigen levels? Transl Androl Urol 2022;11:421-9. [Crossref] [PubMed]
- Van Poppel H, Albreht T, Basu P, et al. Serum PSA-based early detection of prostate cancer in Europe and globally: past, present and future. Nat Rev Urol 2022;19:562-72. [Crossref] [PubMed]
- Stephan C, Vincendeau S, Houlgatte A, et al. Multicenter evaluation of [-2]proprostate-specific antigen and the prostate health index for detecting prostate cancer. Clin Chem 2013;59:306-14. [Crossref] [PubMed]
- Klein EA, Partin A, Lotan Y, et al. Clinical validation of IsoPSA, a single parameter, structure-focused assay for improved detection of prostate cancer: A prospective, multicenter study. Urol Oncol 2022;40:408.e9-408.e18. [Crossref] [PubMed]
- Pinsky PF, Parnes HL, Andriole G. Mortality and complications after prostate biopsy in the Prostate, Lung, Colorectal and Ovarian Cancer Screening (PLCO) trial. BJU Int 2014;113:254-9. [Crossref] [PubMed]
- Kim JY, Yu JH, Sung LH, et al. Usefulness of the prostate health index in predicting the presence and aggressiveness of prostate cancer among Korean men: a prospective observational study. BMC Urol 2021;21:131. [Crossref] [PubMed]
- Lee CU, Lee SM, Chung JH, et al. Clinical Utility of Prostate Health Index for Diagnosis of Prostate Cancer in Patients with PI-RADS 3 Lesions. Cancers (Basel) 2022;14:4174. [Crossref] [PubMed]
- Fossati N, Lazzeri M, Haese A, et al. Clinical performance of serum isoform [-2]proPSA (p2PSA), and its derivatives %p2PSA and the Prostate Health Index, in men aged <60 years: results from a multicentric European study. BJU Int 2015;115:913-20. [Crossref] [PubMed]
- Mottet N, van den Bergh RCN, Briers E, et al. EAU-EANM-ESTRO-ESUR-SIOG Guidelines on Prostate Cancer-2020 Update. Part 1: Screening, Diagnosis, and Local Treatment with Curative Intent. Eur Urol 2021;79:243-62. [Crossref] [PubMed]
- Peters R, Stephan C, Jung K, et al. Comparison of PHI and PHI Density for Prostate Cancer Detection in a Large Retrospective Caucasian Cohort. Urol Int 2022;106:878-83. [Crossref] [PubMed]
- Friedl A, Stangl K, Bauer W, et al. Prostate-specific Antigen Parameters and Prostate Health Index Enhance Prostate Cancer Prediction With the In-bore 3-T Magnetic Resonance Imaging-guided Transrectal Targeted Prostate Biopsy After Negative 12-Core Biopsy. Urology 2017;110:148-53. [Crossref] [PubMed]
- Huang D, Wu YS, Ye DW, et al. Prostate volume does not provide additional predictive value to prostate health index for prostate cancer or clinically significant prostate cancer: results from a multicenter study in China. Asian J Androl 2020;22:539-43. [Crossref] [PubMed]
- DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics 1988;44:837-45. [Crossref] [PubMed]
- Chen H, Qian Y, Wu Y, et al. Modified Prostate Health Index Density Significantly Improves Clinically Significant Prostate Cancer (csPCa) Detection. Front Oncol 2022;12:864111. [Crossref] [PubMed]
- Zhang L, Zhang J, Tang M, et al. MRI-Based Radiomics Nomogram for Predicting Prostate Cancer with Gray-Zone Prostate-Specific Antigen Levels to Reduce Unnecessary Biopsies. Diagnostics (Basel) 2022;12:3005. [Crossref] [PubMed]
- Alijaj N, Pavlovic B, Martel P, et al. Identification of Urine Biomarkers to Improve Eligibility for Prostate Biopsy and Detect High-Grade Prostate Cancer. Cancers (Basel) 2022;14:1135. [Crossref] [PubMed]
- Borghesi M, Ahmed H, Nam R, et al. Complications After Systematic, Random, and Image-guided Prostate Biopsy. Eur Urol 2017;71:353-65. [Crossref] [PubMed]
- Forsvall A, Jönsson H, Wagenius M, et al. Rate and characteristics of infection after transrectal prostate biopsy: a retrospective observational study. Scand J Urol 2021;55:317-23. [Crossref] [PubMed]
- Huang Y, Li ZZ, Huang YL, et al. Value of free/total prostate-specific antigen (f/t PSA) ratios for prostate cancer detection in patients with total serum prostate-specific antigen between 4 and 10 ng/mL: A meta-analysis. Medicine (Baltimore) 2018;97:e0249. [Crossref] [PubMed]
- Chiu ST, Cheng YT, Pu YS, et al. Prostate Health Index Density Outperforms Prostate Health Index in Clinically Significant Prostate Cancer Detection. Front Oncol 2021;11:772182. [Crossref] [PubMed]
- Stephan C, Jung K, Lein M, et al. PHI density prospectively improves prostate cancer detection. World J Urol 2021;39:3273-9. [Crossref] [PubMed]
- Fujita K, Suzuki H, Hinata N, et al. Management of patients with advanced prostate cancer in Japan: 'real-world' consideration of the results from the Advanced Prostate Cancer Consensus Conference. Transl Androl Urol 2022;11:1771-85. [Crossref] [PubMed]
(English Language Editor: A. Muijlwijk)


