
Identification of PDCL2 as a candidate marker in Sertoli cell-only syndrome by chromatin immunoprecipitation-sequencing and bioinformatics analysis
Highlight box
Key findings
• PDCL2 was the most significant downregulated DEG by ChIP-seq in SCOS.
What is known and what is new?
• Understanding the molecular mechanism of SCOS and identifying new non-invasive markers for clinical application is crucial.
• PDCL2 serves as a new biomarker for diagnostic and prognostic value in SCOS.
What is the implication, and what should change now?
• PDCL2 was verified to be a downregulation marker in SCOS, which requires further studies with the biological function and mechanism underlying PDCL2 involvement in SCOS.
Introduction
The male component affects as much as 55% of all infertility couples, and due to the dramatic increase of epigenetic, environmental and lifestyle factors (1,2). According to previously published research, the causes of male infertility mainly include varicocele (3,4), idiopathic or unexplained infertility, hypogonadism, primary testicular failure, endocrine disorders (5,6), obstruction (7,8), cryptorchidism, genetic factors (9,10), anejaculation and retrograde ejaculation and malnutrition (11). The etiology of non-obstructive azoospermia (NOA) encompasses a wide variety of causes, including pre-testicular due to endocrine abnormalities evolving to dysregulation in sex steroids and gonadotropin levels. Other causes include congenital illnesses, like Kallmann syndrome, acquired due to hypothalamic or disorders of the pituitary gland, or even negative feedback or suppression due to exogenous medications or other chemical substances. Testicular causes include congenital illnesses [anorchia, cryptorchidism, Y chromosome microdeletions (YCMD) and other genetic abnormalities], germ cell aplasia [Sertoli cell-only syndrome (SCOS)] and maturation arrest. Acquired testicular causes include trauma, torsion, infection, mumps orchitis and other viral induced damage [i.e., severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)] (12,13), tumors, gonadotoxins, abuse of alcohol and use of other drugs, chemotherapy and radiation therapy, systemic diseases and varicocele.
In SCOS (14), histological diagnosis in testicular biopsies reveals that only Sertoli cells remain in the seminiferous tubules, while the spermatogenic cells are completely absent. SCOS was first described in 1947, but its precise etiology remains largely unknown. Although normal karyotype is found in the majority of SCOS cases, Klinefelter syndrome needs to be ruled out. Clinically, men with NOA and SCOS are likely to present with testicular hypotrophy or even atrophy and abnormal sexual hormones. It is crucial that men obtain at least two semen analyses and proceed to centrifugation of semen samples before initial diagnosis is established (15). Karyotyping and YCMD testing should be routinely performed to assess potential risks to the offspring (16). Differential diagnosis between obstructive azoospermia (OA) and NOA is critical for planning surgical sperm retrieval. The method of choice for addressing NOA patients is usually microdissection testicular sperm extraction (micro-TESE), with overall success rates around 50% (17). However, these invasive operations can result in postoperative complications, including testicular hematoma and inflammation. These complications would cause damage to the few remaining seminiferous ducts, which in turn increases the potential risk of patients being unable to conceive biological offspring or experiencing poor pregnancy outcomes with their spouses. It is critical to develop and validate new non-invasive and more accurate molecular diagnostic markers (18).
Genome-wide transcriptional analysis using gene chips has gained popularity recently as a method for studying disease pathogenesis, disease typing, and identifying the biomarkers of disease development or treatment responses. However, it has had few applications in spermatogenesis to date. Some studies (19-21) have compared the gene expression of normal spermatogenic function, spermatogenic arrest, and SCOS through gene sequencing and indented some genes related to spermatogenesis. However, there is still a significant gap in the number and identification of the critical genes of SCOS.
This study aims to compare the differentially expressed genes (DEGs) in the testicular tissue of patients with SCOS and normal spermatogenesis, and then conducted a bioinformatics analysis to screen and identify the key marker genes. Our findings provide basic data for subsequent research and may extend the understanding of the disease mechanisms of SCOS and provide a nouvelle diagnostic tool to better plan for either clinical and surgical interventions. We present this article in accordance with the MDAR reporting checklist (available at https://tau.amegroups.com/article/view/10.21037/tau-23-304/rc).
Methods
Testis samples
Testis samples was collected from patients with OA and NOA patients at the Department of Urology and Reproductive Center, Peking University Shenzhen Hospital, from October 2021 to September 2022. The inclusion criteria included: (I) microscopic examination without sperm after three consecutive semen centrifugations and (II) requirement for pathological examination of azoospermia and human assisted reproduction. Meanwhile, the exclusion criteria were (I) cryptorchidism, (II) genital and urinary tract infections or severe trauma, (III) non-ejaculation/retrograde ejaculation, (IV) the presence of any karyotype abnormality, including YCMD, 47XXY, 45X, and any mosaicism (V) parotiditis history or varicocele, and (VI) the occurrence of any tumor history, drug and chemotherapy history and radiation therapy history.
Fifty remaining testicular tissues were obtained after pathological examination from 25 SCOS patients and 25 OA patients. OA patients were enrolled to the control group and regarded as normal spermatogenesis. The same pathologist diagnosed all enrolled patients. Micro-TESE and testicular sperm aspiration (TESA) was performed in the volume-dominant testis. The patients were aged between 25 and 45 years and had a mean of 31.0±5.3 years. The testis samples were divided into two parts; one was fixed for pathological examination, and the other was used for further research. This study was approved by the Ethical Committee of Peking University Shenzhen Hospital (No. 202105575). All the patients signed the consent form permitting their samples to be used for scientific research. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Extraction of total RNA and chromatin immunoprecipitation-sequencing (ChIP-seq) experiment
Total RNA was extracted from the fresh testicular tissue using a MiniBEST Universal RNA Extraction Kit (code No. 9767, TaKaRa, Japan). In addition, ultraviolet absorbance spectrophotometry (Olympus, Japan) and denaturing agarose gel electrophoresis (Thermo Fisher, USA) were used to test the purity and integrity of total RNA from the testicular tissue. Arraystar Human messenger RNA (mRNA) Microarray V4.0 (KangCheng, Shanghai, China) was selected for the ChIP-seq experiment. The mRNA data were mainly obtained from the Collaborative Consensus Coding Sequence project.
Image acquisition and gene expression analysis
The Agilent Feature Extraction System (11.0.1.1, Agilent, USA) was used to analyze the array data. GeneSpring GX package (v.12.1, Agilent) was used to normalize and analyze the data processing. The two groups’ statistically significant differentially expressed mRNAs between the two groups were identified based on their P values and fold change (FC) values. P value <0.05 and |logFC| ≥2 were considered as significantly different for DEGs. Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO) analyses were performed to detect the enriched terms and processes of the DEGs. P<0.05 was set as the cut-off criterion (22-24).
Quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA was extracted from the testis samples in the patients with SCOS and the patients with normal spermatogenesis using the MiniBEST Universal RNA Extraction Kit (code No. 9767, TaKaRa) and transcribed into complementary DNA using the Reverse Transcription System (code No. RR037Q, TaKaRa). The sense primer of the main target gene phosducin-like 2 (PDCL2) was (F) 5'-GATGACTCTTGCACAGCTAAAGG-3', and the anti-sense primer was (R) 5'-GAGCTTTCCATTCCTGTAACCGC-3' (synthesized by the Sangon Company, Shanghai, China). The RNA PCR Kit (code No. RR037Q, TaKaRa) was used for the PCR analysis, and each sample was repeated 3 times.
Immunohistochemistry (IHC) analysis
All the testis samples were fixed in Bouin’s solution and embedded in paraffin. The tissues were cut into 2-µm sections, and the antigenicity of the sections was repaired by 0.01 M of sodium citrate buffer (pH 6.0) for 3 min. Polyclonal antibody PDCL2 (dilution: 1:200, RRID: AB_2299492, catalogue number: 17407-1-AP, Proteintech, USA) was hatched at room temperature for 2 hours, and subsequently incubated with the second antibody for 30 min. Next, the signal of the sections was developed by diaminobenzidine, and hematoxylin was used to counterstain the sections. Brown granules indicated positive results. Light microscopy (Olympus BX43) was used for the visualization.
Statistical analysis
All the data were analyzed by SPSS 28.0 (IBM Corp, Armonk, NY, USA). The parameters of two groups conforming to the normal distribution were compared by t-test. A P value <0.05 indicated that the difference was statistically significant.
Results
Identification of the DEGs in SCOS
The samples collected from 5 patients with SCOS and 5 patients with normal spermatogenesis were used for the RNA-sequencing (RNA-seq) experiments. All the samples passed the purity test and quality requirements (A260/A280 between 1.81 and 1.97). A total of 15,961 genes were screened out by the gene ChIP-seq, including 4,539 significant DEGs (P<0.05), of which 3,133 were upregulated and 1,406 were downregulated (Figure 1A-1C, Table 1).
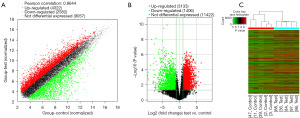
Table 1
| DEGs | Gene names |
|---|---|
| Upregulated genes | NPPB, HSD17B3, ARX, HSD3B2, PDC, AUTS2, SULT2A1, TEK, STAR, C21orf62, PTPRO, CYP11A1, GP5, MYL7, HBA1, HBB, MUM1L1, GSTA3, PCDHB17, AP000783.1, BZRAP1, MS4A12, HEG1, LOC440243, ACTN2, C6, TRIM6-TRIM34, HBG2, SYT2, FAM133A, HHIP, C8orf12, LINGO2, TREM2, ALDH1A3, NXF5, HS6ST2, PLAC1, SLC4A3, TNNT3, NAT2, TRPM3, REP15, CT47A1, WIPF3, DPEP1, HSD3B1, UTY, TCF21, KCNC3, EYA4, KRT32, HBA2, C7, CGB2, IGFBP6, ITLN1, SLC39A11, LAG3, GATA5, NUDT10, SDIM1, CBLN4, NXF2B, SMAD9, MYO7A, SOX3, SLC51A, ITLN2, PLA2G5, PON1, TMEFF2, SMN2, DRD5, KCNIP3, DIO3, ZNF502, TRIM55, RNF207, AGRP, PHF24, ST6GALNAC1LHX9, AK123878, B3GALT2, MTCP1, TCEAL2, PRPS2, MRO, KLHL4, TNKS1BP1, GADD45G, SEPT14, CLEC4M, METAP1D, COLEC11, ANKDD1A, SMIM1, AL117190.3, SPINK13 |
| Downregulated genes | PDCL2, CAPZA3, KRTDAP, SPACA4, KRT15, ESRP2, XAGE1C, CCDC158, LDHC, KDF1, GTSF1, UBL4B, C16orf82, TP53TG5, KRT23, BNC1, MFSD6L, PROK2, TTC29, LGALS7, TNP1, ROPN1L, KRT5, DKKL1, CRISP2, C2orf57, CCNB2, RBBP8NL, CALML5, C12orf56, SDR16C5, ROPN1B, SHCBP1L, BC016978, PRSS37, UBE2U, CERS3, WDR63, SCGB2A2, TSACC, CCIN, CRISP3, GPR18, HORMAD1, GJB6, NCAPG, SPINK2, FSIP2, CMTM2, GPRIN2, TJP3, ACTRT2, SPATA8, TIMD4, NUPR1L, HOXA11, STARD6, CDKN3, SLC22A23, TEX40, CDK1, DNAH3, ROPN1, TYRP1, PPP1R36, CDRT15L2, PRSS21, C5orf58, RSPH6A, RPL10L, PBK, FTMT, MLF1IP, SPATA32, KRT6B, CLCA2, SAA2, LGALS7B, DEPDC1B, PIWIL1, MYRFL, TM4SF19, MFAP3L, BTBD16, ADGB, STMN2, ART3, CATSPERB, PP2D1, PRAC2, SPATA3, PHF7, CDC45, ANKRD30A, SLPI, GAGE4, MSMB, CATSPERD, LOC100505841, SMCP |
DEGs, differentially expressed genes; SCOS, Sertoli cell-only syndrome.
GO term analysis of the DEGs in SCOS
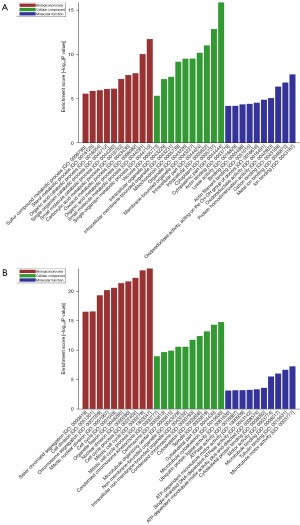
A GO analysis was performed on the DEGs using a cut-off value of P<0.05. In the biological process (BP) domain, the upregulated DEGs were mainly enriched in the single-organism and small-molecule metabolic processes. In the cellular component (CC) domain, the top 3 processes of the upregulated DEGs were the cytoplasmic part, cytoplasm, and intracellular. In the molecular functional (MF) domain, the highest-ranked processes of the upregulated DEGs were ion binding, metal-ion binding, and cation binding (Figure 2A). Meanwhile, in the BP domain, the downregulated DEGs were mainly enriched in the mitotic, mitotic, and cell cycle processes. In the CC domain, the top 3 processes of the downregulated DEGs were microtubule binding, the cytoskeletal part, and the spindle. In the MF domain, the highest-ranked processes of the downregulated DEGs were microtubule motor activity, tubulin binding, and microtubule binding (Figure 2B).
KEGG pathway enrichment analysis in SCOS
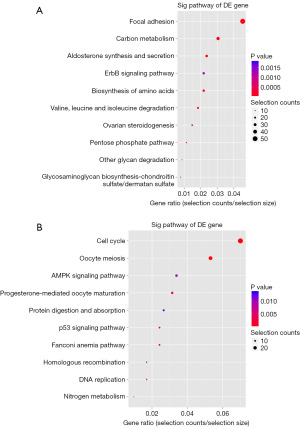
A KEGG pathway analysis was performed on the DEGs using a cut-off value of P<0.05. In total, 71 pathways were regulated by the upregulated mRNAs in the testicular tissue, of which the most enriched pathways were valine, leucine, and isoleucine degradation, which consisted of 21 DEGs (Figure 3A). A total of 22 pathways were regulated by the downregulated mRNAs, of which the most enriched pathway was the cell cycle, which consisted of 29 DEGs (Figure 3B). Among the downregulated mRNAs, 22 DEGs were enriched in the oocyte meiosis pathway, and 7 DEGs were enriched in the homologous recombination pathway. There were more downregulated DEGs than upregulated DEGs involved in spermatogenesis.
Expression of PDCL2 in SCOS
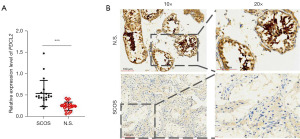
Based on the ChIP-seq results, bioinformatics analysis and literature reports, we selected the mRNA PDCL2 as the target gene for further research, as it had the most considerable fold difference among the downregulated DEGs and restricted expression in the testis. qRT-PCR and IHC analyses were performed to verify the expression of PDCL2 between the testis samples of the 20 patients with SCOS and the 20 patients with normal spermatogenesis. The qRT-PCR results showed that the expression of PDCL2 was significantly decreased (P<0.001) in the SCOS group compared to the normal spermatogenesis group (Figure 4A). The IHC results showed that PDCL2 was highly expressed in the germ cells cytoplasm in the human testis, and PDCL2 expression was higher in the normal spermatogenesis group than in the SCOS group (Figure 4B).
Discussion
Only a few studies (25-27) have systematically sequenced the different genes of SCOS. The physiopathological misunderstanding surrounding a significant part of patients presenting with azoospermia results in the wrongful clinical evaluation of these individuals, leading to biased, confused, and often insufficient data gathering and interpretation. Therefore, these critical problems may hinder basic research and the clinical management of patients presenting with SCOS. In this study, we used gene RNA-seq to screen the DEGs in the testicular tissue of patients with SCOS and normal spermatogenesis. In total, 4,539 significant DEGs were obtained, of which 3,133 were upregulated, and 1,406 were downregulated. About bioinformatics analysis, the mitotic cell cycle process, single-organism metabolic process, cytoplasmic part, microtubule binding, microtubule motor activity, and CC were the top terms of the GO analysis. In contrast, valine, leucine and isoleucine degradation, carbon metabolism and the cell cycle were the top processes of the KEGG analysis. Notably, oocyte meiosis, Fanconi’s anemia DNA replication, and the oocyte maturation pathways involved in spermatogenesis regulation were also highly enriched.
The pathological feature of SCOS is the complete apoptosis of germ cells in the seminiferous tubules (14). Thus, this study focused on the cell cycle, proliferation, and apoptosis pathways. As PDCL2 had the most significant fold difference among the downregulated DEGs, had restricted expression in the testis, and was mainly enriched in the cell cycle, PDCL2 was chosen as the target gene for further research. PDCL2 is located on chromosome 4 in humans and was found to be mainly expressed in male and female germ cells (28). Li et al. (29) found that PDCL2 protein is a membrane protein that plays an essential role in the maturation of germ cells. The apoptosis of testicular germ cells in mice was significantly increased after PDCL2 knockout. The possible mechanism of PDCL2 may be related to the interaction of the cytosolic chaperonin-containing T-complex polypeptide 1 (CCT) complex and the dysfunction of this complex. Fujihara et al. (30) found that PDCL2-deficient sperm had a round head. Observations of the testes of PDCL2-knockout mice revealed the loss of sperm acrosomal cap formation and malformed nuclei. It has also been reported that the PDCL2 protein is associated with the decreased survival of kidney cancer patients (31).
We found that PDCL2 was mainly expressed in the cytoplasm of the germ cells in humans. The qRT-PCR and IHC results showed that the expression of PDCL2 was significantly downregulated in the testis of the SCOS patients. These results suggest that PDCL2 may be involved in the process of SCOS and the regulation of apoptosis in germ cells. PDCL2 may serve as a new biomarker for diagnostic and prognostic value in individuals presenting with SCOS in order to facilitate surgical planning (TESA or micro-TESE) with more realistic successful rates or even to avoid unnecessary procedures that could eventually lead to hormonal deficiency and hypogonadism in particular in those cases with a first or multiple failed retrieval attempts. The biological function and mechanism underlying PDCL2 involvement in SCOS has been hitherto uncovered here. Nevertheless, it undoubtedly and desirably requires further studies with different SCOS populations and genetic backgrounds.
This study provided valuable data that robustly examined one new aspect of the molecular mechanisms behind SCOS and provided a new investigative and diagnostic tool to optimize laboratory and clinical investigation. Besides, we could provide patients with SCOS with more information on the likelihood of finding spermatozoa in procurement procedures, optimizing time, financial and emotional costs, stress, and surgical complications.
Conclusions
The testicular samples in this study originated from patients with SCOS and normal spermatogenesis, providing realistic data from the desired study-populations. In total, 3,133 upregulated DEGs and 1,406 downregulated DEGs were obtained by the RNA-seq. The mitotic cell cycle, cell cycle, and oocyte maturation were highly enriched processes involved in spermatogenesis. In addition, PDCL2 was verified to be a downregulation marker by qRT-PCR and IHC in SCOS. These findings may extend the understanding of the processes and mechanism of SCOS and contribute to the development of diagnostic tools, more robust and precise clinical and therapeutical management of patients presenting with SCOS.
Acknowledgments
Funding: This work was supported by a grant from the Project of Administration of Traditional Chinese Medicine of Guangdong, China (No. 20221014).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://tau.amegroups.com/article/view/10.21037/tau-23-304/rc
Data Sharing Statement: Available at https://tau.amegroups.com/article/view/10.21037/tau-23-304/dss
Peer Review File: Available at https://tau.amegroups.com/article/view/10.21037/tau-23-304/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tau.amegroups.com/article/view/10.21037/tau-23-304/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the Ethical Committee of Peking University Shenzhen Hospital (No. 202105575). All the patients signed the consent form permitting their samples to be used for scientific research.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Agarwal A, Mulgund A, Hamada A, et al. A unique view on male infertility around the globe. Reprod Biol Endocrinol 2015;13:37. [Crossref] [PubMed]
- Pan MM, Hockenberry MS, Kirby EW, et al. Male Infertility Diagnosis and Treatment in the Era of In Vitro Fertilization and Intracytoplasmic Sperm Injection. Med Clin North Am 2018;102:337-47. [Crossref] [PubMed]
- Gat I, Madgar I. The varicocele enigma: "background noise" or common male infertility etiology? Harefuah 2015;154:312-5, 338-9. [PubMed]
- Huang HC, Huang ST, Chen Y, et al. Prognostic factors for successful varicocelectomy to treat varicocele-associated male infertility. Reprod Fertil Dev 2014;26:485-90. [Crossref] [PubMed]
- Santi D, Granata AR, Simoni M. FSH treatment of male idiopathic infertility improves pregnancy rate: a meta-analysis. Endocr Connect 2015;4:R46-58. [Crossref] [PubMed]
- Lei B, Lv D, Zhou X, et al. Biochemical hormone parameters in seminal and blood plasma samples correlate with histopathologic properties of testicular biopsy in azoospermic patients. Urology 2015;85:1074-8. [Crossref] [PubMed]
- Taniguchi H, Iwamoto T, Ichikawa T, et al. Contemporary outcomes of seminal tract re-anastomoses for obstructive azoospermia: a nationwide Japanese survey. Int J Urol 2015;22:213-8. [Crossref] [PubMed]
- van Wely M, Barbey N, Meissner A, et al. Live birth rates after MESA or TESE in men with obstructive azoospermia: is there a difference? Hum Reprod 2015;30:761-6. [Crossref] [PubMed]
- Li LL, Peng D, Wang RX, et al. Correlation between chromosomal polymorphisms and male infertility in a Northeast Chinese population. Genet Mol Res 2015;14:15435-43. [Crossref] [PubMed]
- Richardson ME, Bleiziffer A, Tüttelmann F, et al. Epigenetic regulation of the RHOX homeobox gene cluster and its association with human male infertility. Hum Mol Genet 2014;23:12-23. [Crossref] [PubMed]
- Giahi L, Mohammadmoradi S, Javidan A, et al. Nutritional modifications in male infertility: a systematic review covering 2 decades. Nutr Rev 2016;74:118-30. [Crossref] [PubMed]
- Teixeira TA, Oliveira YC, Bernardes FS, et al. Viral infections and implications for male reproductive health. Asian J Androl 2021;23:335-47. [Crossref] [PubMed]
- Duarte-Neto AN, Teixeira TA, Caldini EG, et al. Testicular pathology in fatal COVID-19: A descriptive autopsy study. Andrology 2022;10:13-23. [Crossref] [PubMed]
- Nagpal BL, Manjari M, Kapoor K, et al. Testicular biopsy in cases of male infertility: a retrospective study. J Indian Med Assoc 1993;91:171-4. [PubMed]
- Monteiro RA, Pariz JR, Pieri Pde C, et al. An easy, reproducible and cost-effective method for andrologists to improve the laboratory diagnosis of nonobstructive azoospermia: a novel microcentrifugation technique. Int Braz J Urol 2016;42:132-8. [Crossref] [PubMed]
- Hallak J. A call for more responsible use of Assisted Reproductive Technologies (ARTs) in male infertility: the hidden consequences of abuse, lack of andrological investigation and inaction. Transl Androl Urol 2017;6:997-1004. [Crossref] [PubMed]
- Schlegel PN, Li PS. Microdissection TESE: sperm retrieval in non-obstructive azoospermia. Hum Reprod Update 1998;4:439. [Crossref] [PubMed]
- McLachlan RI, Rajpert-De Meyts E, Hoei-Hansen CE, et al. Histological evaluation of the human testis--approaches to optimizing the clinical value of the assessment: mini review. Hum Reprod 2007;22:2-16. [Crossref] [PubMed]
- Wang M, Liu X, Chang G, et al. Single-Cell RNA Sequencing Analysis Reveals Sequential Cell Fate Transition during Human Spermatogenesis. Cell Stem Cell 2018;23:599-614.e4. [Crossref] [PubMed]
- Wang Z, Xu X, Li JL, et al. Sertoli cell-only phenotype and scRNA-seq define PRAMEF12 as a factor essential for spermatogenesis in mice. Nat Commun 2019;10:5196. [Crossref] [PubMed]
- Lin YH, Lin YM, Teng YN, et al. Identification of ten novel genes involved in human spermatogenesis by microarray analysis of testicular tissue. Fertil Steril 2006;86:1650-8. [Crossref] [PubMed]
- Faghihi MA, Wahlestedt C. Regulatory roles of natural antisense transcripts. Nat Rev Mol Cell Biol 2009;10:637-43. [Crossref] [PubMed]
- Ørom UA, Derrien T, Beringer M, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell 2010;143:46-58. [Crossref] [PubMed]
- Cabili MN, Trapnell C, Goff L, et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev 2011;25:1915-27. [Crossref] [PubMed]
- Shiraishi K, Tabara M, Matsuyama H. Transcriptome Analysis to Identify Human Spermatogonial Cells from Sertoli Cell-Only Testes. J Urol 2020;203:809-16. [Crossref] [PubMed]
- Wang X, Li Z, Qu M, et al. A homozygous PIWIL2 frameshift variant affects the formation and maintenance of human-induced pluripotent stem cell-derived spermatogonial stem cells and causes Sertoli cell-only syndrome. Stem Cell Res Ther 2022;13:480. [Crossref] [PubMed]
- Maatouk DM, Natarajan A, Shibata Y, et al. Genome-wide identification of regulatory elements in Sertoli cells. Development 2017;144:720-30. [Crossref] [PubMed]
- Aisha J, Yenugu S. Characterization of SPINK2, SPACA7 and PDCL2: Effect of immunization on fecundity, sperm function and testicular transcriptome. Reprod Biol 2023;23:100711. [Crossref] [PubMed]
- Li M, Chen Y, Ou J, et al. PDCL2 is essential for spermiogenesis and male fertility in mice. Cell Death Discov 2022;8:419. [Crossref] [PubMed]
- Fujihara Y, Kobayashi K, Abbasi F, et al. PDCL2 is essential for sperm acrosome formation and male fertility in mice. Andrology 2023;11:789-98. [Crossref] [PubMed]
- Jamin SP, Hikmet F, Mathieu R, et al. Combined RNA/tissue profiling identifies novel Cancer/testis genes. Mol Oncol 2021;15:3003-23. [Crossref] [PubMed]


