
Clinical and molecular features of 40 Chinese patients with idiopathic hypogonadotropic hypogonadism
Highlight box
Key findings
• Ten new genetic mutations related to IHH in four families and eight sporadic unrelated IHH patients were identified.
What is known and what is new?
• The incidence of IHH is 1–10:100,000; about 60% of patients with IHH have complete or partial loss of smell (KS), and the ratio of men to women is as high as 5:1.
• The total positive detection rate of 40 patients was 30% (nIHH 8/18 + KS 4/22), and the FGFR1 mutation rate accounted for 7.5% (3/40). Mutation rates of ANOS1, CHD7, and KISS1R were 5% (2/40), respectively. The mutation rates of SEMA3E, PROKR2, and SOX10 were 2.5% (1/40), respectively.
What is the implication, and what should change now?
• The study aims to expand the genotype mutation spectrum of IHH and provide evidence for the follow-up clinical treatment and genetic counseling of the disease.
Introduction
Idiopathic/isolated hypogonadotropic hypogonadism (IHH) is a rare disease caused by impaired congenital hypothalamic gonadotropin-releasing hormone (GnRH) neuronal function, impaired synthesis, secretion, or function of GnRH results in decreased secretion of gonadotropin by the pituitary gland, also known as congenital hypogonadotropic hypogonadism (CHH) (1). Delayed puberty is the most typical manifestation of IHH disease is a clinical and genetically heterogeneous disease with sporadic and familial characteristics. It has been reported that a single gene mutation does not only cause IHH disease; some are also caused by multiple gene mutations (2). At present, the known gene mutations can only explain 30–50% of the pathogenic causes, and there are still primarily unknown pathogenic genes and related pathogenic mechanisms to be explored.
Slight and transient reactivation of the Hypothalamus-Pituitary-Gonad (HPG) axis in early infancy (approximately 4 to 16 weeks) provides a unique opportunity to diagnose congenital IHH in both male and female infants (3). GnRH neuron originates from the basal olfactory plate of the olfactory system and moves to the hypothalamus. The hypothalamus secretes GnRH, which is transported to the anterior pituitary through the capillary network, stimulating follicle-stimulating hormone (FSH) and luteinizing hormone (LH) secretion to control reproduction (4). Traditionally, congenital IHH can be divided into two categories: (I) Kallmann syndrome (KS) is caused by developmental defects of GnRH neurons and olfactory bulbs, with varying degrees of loss or reduction of smell (5); (II) nIHH is usually caused by lack of FSH and LH due to GnRH secretion or interruption of function (6). The incidence of IHH is 1–10:100,000; about 60% of patients with IHH have complete or partial loss of smell (KS), and the ratio of men to women is as high as 5:1 (7). In addition, compared with affected female patients, IHH male patients show a more severe phenotype in the family (1).
Applying new technologies such as whole exome sequencing (WES) has increased the molecular diagnosis rate of IHH patients from approximately 30% to 50% (8). So far, the genes closely related to the molecular pathogenesis of the disease are ANOS1 (KAL1), LHB, FSHB, GNRHR, FGFR1, KISS1R, SOX2, PROKR2, CHD7, FGF8, GNRH1, KISS1, SEMA3A, FGF17, SOX10, DMXL2, SEMA3E, AMH, AMHR2, SPRY4, SEMA7A, etc. The proteins encoded by these genes are involved in developing and migrating GnRH neurons or controlling different stages of GnRH function (9). It has been reported in the literature that mutations in genes related to GnRH secretion (GNRH1, GNRHR, KISS1/KISS1R, TAC3/TACR3, etc.) generally cause nIHH (6).
To better understand the molecular genetic basis of IHH, we retrospectively recorded 40 unrelated IHH patients for a series of Sanger sequence, WES, and bioinformatics analyses. Our data provide an essential foundation for patients’ follow-up clinical diagnosis, treatment, and genetic counseling and expand the disease’s genotype-phenotype spectrum. We present this article in accordance with the MDAR reporting checklist (available at https://tau.amegroups.com/article/view/10.21037/tau-23-225/rc).
Methods
Clinical research of samples
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by ethics board of the Jinling Hospital, Nanjing University School of Medicine (No. 2018NZGKJ-003) and informed consent was taken from the patients or legal guardians. We diagnosed 40 IHH patients (22 KS and 18 nIHH) from 2014 to 2021 in Nanjing Jinling Hospital. All the patients were Han Chinese males who came to our hospital for gonadal dysplasia or fertility problems.
Sample collection criteria: (I) reproductive phenotype: small penis, small testis or cryptorchidism or penis testis were childish, secondary sex characteristics were not prominent, etc. Patients with or without anosmia; a family history of related diseases; (II) sex hormone tests: low testosterone (T), very low FSH and LH levels or below the upper limit of normal [(in general: T <3.47 nmol/L, LH <0.7 IU/L), while prolactin (PRL) values were average (to exclude Prolactinoma)]; (III) brain MRL/CT showing normal or underdeveloped olfactory lamina and/or olfactory sulci (to exclude pituitary tumors); (IV) chromosome karyotype 46, XY without Y chromosome microdeletion; (V) other related hormone tests showed no abnormalities.
We screened olfactory using 75% medical alcohol, distilled water, and acetic acid; abnormal olfactory was defined as KS and normal olfactory was nIHH. We examined the age and weight of the patients, surgical history, PRL, FSH, LH, and T. We assessed the left and right testicular volumes and performed semen density analysis according to WHO criteria.
We collected the peripheral blood of patients and some family members to extract genomic DNA (gDNA) from blood. The quality of DNA samples was tested as follows: the total amount of DNA samples should be ≥200 ng, and the concentration should be ≥40 ng/µL. We detected the purity of DNA samples by NanoPhotometerR spectrophotometer, and Qubit 8.3.0 Fluorometer was utilized to detect the concentration of DNA samples.
WES, Sanger sequencing, and bioinformatics analysis
We performed the WES to 40 IHH probands, and the sequencing company assisted with WES. WES experiments for this study were conducted with the assistance of Beijing Nuohe Zhiyuan BioInformation Technology Co., Ltd. Specific steps: first, we extracted 3mL of peripheral blood from all participants to extract gDNA. We tested the sample quality again; the total amount of DNA was ≥2 ng, and the concentration was ≥40 ng/µL. Then the library construction and quality inspection. Then the samples were sequenced on the machine, and we analyzed the sequencing results informatically.
We verified all gene rare sequencing variants (RSVs) by Sanger sequencing. In only four patients, we conducted the pedigree investigation and related clinical examination for objective or family reasons. We designed primer using primer 5.0 software and amplified the exome regions of possible disease gene mutation sites analyzed by the proband and some families from the WES results.
We verified the success of polymerase chain reaction (PCR) by agarose gel electrophoresis and the gel imaging system. If the position of the PCR product band coincides with the target fragment, we performed the Sanger sequencing map with Chromas software. Check whether the peak map of the mutation site of the proband is consistent with that of the control group, and if not, verify whether the sequencing result is consistent with the WES consequence. Then, observe whether the family members also appear in this phenomenon, analyze whether it is compatible with the sense of co-separation of the mutation site and the disease, and finally, query whether the mutation site is novel.
Mutations detected in the study are based on the rules for naming mutations established by the Human Genome Variation Society (HGVS). According to the American College of Medical Genetics and Genomics (ACMG) guidelines, the biological information of rare mutation sites, including the correlation of mutations and diseases, mutation pathogenicity, minimum allele frequency of the human genome database, etc. We utilized MEGA5 software to analyze the homology of amino acids encoded by gene mutation sites between different species to determine this mutation site’s evolutionary conservation. The variants are separated into five categories: pathogenic, possibly pathogenic, of unknown significance, perhaps benign, and benign. In addition, based on SIFT and PolyPhen-2 software to classify missense mutations, if the mutation site is likely to be harmful by SIFT or PolyPhen-2, this variant can be classified as pathogenic (10,11). Based on mutation Taster software analysis of nonsense mutations, if the prediction is harmful, this variant can be classified as pathogenic (12). Because their highly destructive effects on protein structure or expression are pathogenic, frameshift mutations have not been confirmed further.
Statistical analysis
The data were analyzed using SPSS software (version 23.0; IBM Corp., Armonk, NY, USA). Mean values were compared between groups using the Maine-Whitney U test for normally distributed data and the Wilcoxon test for paired data. A P value <0.05 was considered statistically significant.
Results
Clinical data of all patients
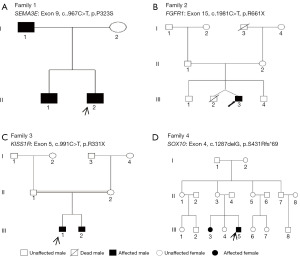
Our routine clinical examination of 4 patients with family history and 8 patients with sporadic IHH found that all patients had low hormone levels. Probands 1, 4, 5, and 6 were associated with olfactory disorders, and all the remaining patients had the usual olfactory sense. Patient 4 examined non-reproductive phenotype with sensorineural hearing loss and Iris heterochromatic. The clinical characteristics of patients carrying disease-causing genes are described in Table 1. We carried out a pedigree investigation on four patients (Figure 1). Proband 1’s father and brother had hyposmia and were diagnosed with KS. We examined the brother of proband 3 with an abnormal penis, left and right testicular volume 2 mL, and diagnosed with nIHH. The sister of proband 4 was concerned by sensorineural hearing and olfactory impairment.
Table 1
| Pt. | Age at diagnosis (years) | Clinical diagnosis | Olfaction | Reproductive phenotype | Basal hormone levels (T nmol/L; FSH IU/L; LH IU/L) | Mutation | Hem | SIFT_score/pred | Polyphen2_HVAR_score/pred | MutationTaster_score/pred |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 29 | KS | Anosmia | Penis: 5 cm, testicular volume: left and right sides 2 mL | T 0.54; FSH 0.69; LH 0.15 | NG_021242.2: c.967C>T (SEMA3E: Exon9, c.967C>T, p.P323S) | 1 | 0.005/D | 0.998/D | 1/D |
| 2 | 16 | nIHH | Normal | External genital dysplasia | T 0.18; FSH 0.2; LH 0.3 | NG_007729.1: c.1981C>T (FGFR1: Exon15, c.1981C>T, p.R661X) | 1 | / | / | 1/A |
| 3 | 26 | nIHH | Normal | Penis abnormal, testicular volume: left and right sides 5 mL | T 0.2; FSH 1.67; LH 0.67 | NG_008277.1: c.991C>T (KISS1R: Exon5, c.991C>T, p.R331X) (reported) | 0 | / | / | 1/A |
| 4 | 22 | KS | Anosmia | Delayed puberty, penile, testicular dysplasia | T 0.69; FSH 0.4; LH2.2 | NG_007948.1: c.1287delG (SOX10: Exon4, c.1287delG, p.S431Rfs*69) | 1 | / | / | / |
| 5 | 31 | KS | Hyposmia | Penis:8 cm, testicular volume: left and right sides 2 mL | T 1.14; FSH 1.6; LH 1.1 | NG_007088.2: c.844delC (ANOS1: Exon6, c.844delC, p.R282Vfs*26) | 0 | / | / | / |
| 6 | 25 | KS | Anosmia | Penis normal, testicular volume: left 2 mL, right 3 mL, had history of cryptorchidism | T 23; FSH 1.45; LH 0.45 | NG_007088.2: c.1886_1887insCTACTCT (ANOS1: Exon13, c.1886_1887insCTACTCT, p.R631Sfs*36) | 0 | / | / | / |
| 7 | 27 | nIHH | Normal | Left testicular volume 2 mL, right cryptorchidism | T 5.86; FSH 0.51; LH 0.37 | NG_007729.1: c.2117_2118insT (FGFR1: Exon16, c.2117_2118insT, p.E705Gfs*16) | 1 | / | / | / |
| 8 | 27 | nIHH | Normal | Left testicular volume 1 mL, right cryptorchidism | T 0.55; FSH 1.84; LH 0.44 | NG_007729.1: c.2239_2255delTTCAAGCAGCTGGTGGA (FGFR1: Exon17, c.2239_2255delTTCAAGCAGCTGGTGGA, p.F745Rfs*45) | 1 | / | / | / |
| 9 | 23 | nIHH | Normal | Penis abnormal, left and right testicular volume: abnormal | T 0.59; FSH 0.1; LH 0.1 | NG_007009.1: c.5695_5698delGGCC | 1 | / | / | / |
| NG_007009.1: c.5700_5707delACTTTACT (CHD7: Exon29, c.5695_5698delGGCC, c.5700_5707delACTTT ACT, p.G1899R, Q1900_W1903del) | ||||||||||
| 10 | 26 | nIHH | Normal | Penis normal, testicular volume: left 2 mL, right 3 mL, right had history of cryptorchidism | T 2.87; FSH 0.2; LH 0.6 | NG_007009.1: c.5355G>T (CHD7: Exon25, c.5355G>T, p.W1785C) | 1 | 0/D | 0.993/D | 1/D |
| 11 | 30 | nIHH | Normal | Left and right testicular volume: 6–8 mL | T 1.52; FSH 1.7; LH 1.3 | NG_008132.2: c.667T>G | 1 | 0/D | 1/D | 1/D |
| NG_008132.2: c.892C>T (PROKR2: Exon2, c.667T>G, p.Y223D c.892C>T, p.R298C) | ||||||||||
| 12 | 24 | nIHH | Normal | Penis normal, left and right testicular volume: 1–2 mL | T 0.76; FSH 0.98; LH <0.1 | NG_008277.1: c.309C>A (KISS1R: Exon2, c.309C>A, p.Y103X) (reported) | 0 | / | / | 1/A |
The testicular volume of Chinese men is about 15–25 mL. Normal levels of male sex hormone: T, 9.4–37 nmol/L; FSH, 1.2–5.5 IU/L; LH, 1.3–6.3 IU/L. Hem_1: heterozygous mutation; Hem_0: homogeneous mutation; A: disease_causing_automatic; D: disease_causing. IHH, idiopathic hypogonadotropic hypogonadism; T, testosterone; FSH, follicle-stimulating hormone; LH, luteinizing hormone; KS, Kallmann syndrome; nIHH, normal IHH.
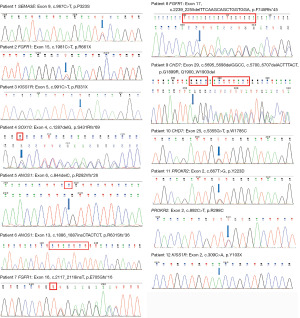
WES and Sanger sequencing results of all patients
We filtered the WES data and analyzed it to identify important mutation sites. WES results showed that 12 cases (4 families and 8 sporadic cases) carried gene mutations. Sanger sequencing was consistent with WES results; members of four families were sequenced by Sanger sequencing (Table S1), confirming that the mutated sites were co-isolated with genotype and disease (Figure 2 and Figure 3). Two patients carried the known mutation site of KISS1R. The remaining 10 patients took new mutation sites of knowing disease-causing genes. We determined that a gene mutation was the leading cause of IHH in 4 cases of pedigree and 8 cases of sporadic IHH patients. Four mutations were found to cause IHH in the corresponding pedigree. Except for KISS1R, the remaining mutations are absent in the Human Gene Mutation Database, HPSD, and the dbSNP.
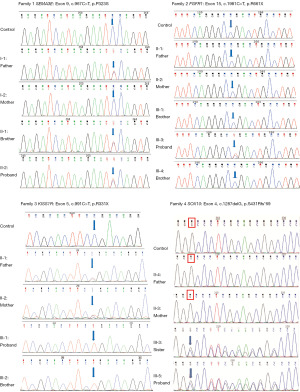
Bioinformatics and mutation rate analysis
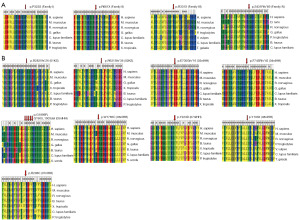
The overall positive detection rate of 40 patients was 30% (nIHH 8/18 + KS 4/22), with the FGFR1 mutation rate accounting for 7.5% (3/40). ANOS1, CHD7, and KISS1R mutation rates were 5% (2/40), respectively. The mutation rate of SEMA3E, PROKR2, and SOX10 was 2.5% (1/40), respectively. We found that all missense mutation sites SEMA3E (p.P323S), CHD7 (p.W1785C), PROKR2 (p.Y223D and p.R298C) were harmful; all nonsense mutation sites FGFR1 (p.R661X), KISS1R (p.R331X, p.Y103X) are pathogenic (Table 1). The comparison of MEGA5 software shows that all the variants have extremely high homology among different species and are strongly conservative in evolution (Figure 4).
Discussion
SEMA3E gene mutations are primarily related to KS and affect the survival of GnRH neurons. Cariboni et al. (13) demonstrated that during hypothalamic migration, SEMA3E and the receptor PLXND1 are essential factors for the survival of GnRH neurons in brain development. In addition, through WES and computer model analysis, SEMA3E (p.R619C) and CHD7 (p.F1019C) mutations were identified among the two brothers with KS (14). The specific R619C missense mutation caused selective death and apoptosis in central GnRH neurons. Both mutations are inherited from the parents, and we studied the family I patients in this chapter similarly. It further illustrates the clinical heterogeneity of KS caused by SEMA3E gene mutation.
FGFR1, a loss of function mutation in approximately 10% of patients with IHH, is also required for the migration, differentiation, survival, and secretion of normal GnRH neurons (8). Xu et al. (15) reported a case of a 2.5 years old male patient with low serum hormones (T, FSH, LH, etc.), cryptorchidism, and normal olfaction. Genetic testing revealed that the boy carried a novel heterozygous mutation of FGFR1 (c.1889T>C, p.L630P). In family Ⅱ, only the proband carries the FGFR1, c.1981C>T mutation, which is a new mutation and is not inherited from the parents. We predict that the proband FGFR1, p.R661X, may also disrupt FGF8/FGFR1 signaling, the main pathway for GnRH neuron development (8,16). In 07 and 08 nIHH patients, the amino acid residues of E707 and F747 are also located in the highly conserved tyrosine kinase domain of FGFR1, which plays a catalytic role. We predict that FGFR1, p.E705Gfs*16, and p.F745Rfs*45 will decrease tyrosine kinase activity, resulting in downstream MAPK signaling failure and affecting GnRH neuron secretion.
Seminara et al. (17) reported that one IHH patient carried KISS1R p.R331X and p.X399R mutations. The KISS1R mutation causes hypogonadism and delays puberty development in humans and mice, which can be improved by administering exogenous GnRH (18). Both brothers in Family III have nIHH disease, and the parents are consanguineous in marriage. Both the proband and the younger brother are homozygous carriers of known KISS1R, c.991C>T mutation, and both parents carry the heterozygous mutation site of the gene. The 12th nIHH patient carries a known homozygous mutation of KISS1R, c.309C> A, p. Y103X. Y103X completely switched the receptor’s structure by deleting five transmembrane domains, and MAPK/ ERK/SRE luciferase detection did not respond (19).
SOX10 contains a highly conserved HMG (high mobility group) structure that can precisely determine and bind DNA sequences domain (20). According to a literature review, SOX10 mutations account for 4.4% of IHH patients studied (21). SOX10 is an essential transcription factor during neural crest development and can interact with other proteins (such as PAX3) to regulate neural crest cell development (22,23). The transcription factor SOX10 is an essential regulator of the neural crest and the central nervous system (CNS) in all vertebrates, so mutation or deletion of the SOX10 gene leads to severe developmental disorders (20).
For unrelated men with sporadic KS, the incidence of ANOS1 mutations may range from 3.5% to 10% (24,25). In sporadic unrelated IHH patients 05 and 06, R282 and L629 amino acid residues are located in or near the fibronectin type III domain. We predict that ANOS1 mutation in these 2 KS patients will cause defects in kinase activity, likely disrupting neuronal migration and axon orientation (26).
About 6% of IHH patients (with or without olfactory impairments) had the CHD7 mutation (7). CHD7 mutation is also the primary pathogenic gene causing CHARGE syndrome, so we need to explore further the CHARGE syndrome in patients 09 and 10 (27). Recently, a study has demonstrated that CHD7 is pivotal in developing GnRH neurons and the olfactory system (28). Studies have revealed that PROKR2 knockout gene mice exhibit dysplasia of the olfactory bulb, and migration of GnRH neurons fails (29). Functional loss mutations in PROKR2 have been reported in approximately 5.6% of IHH patients (30). The 11th nIHH patient carries both c.667T>G and c.892C>T mutations on PROKR2.
In a clinical sense, recognizing those IHH genes and associated phenotypes may improve our diagnostic capabilities by enabling us to prioritize the screening of particular genes such as synkinesia (ANOS1), dental agenesis (FGF8/FGFR1) and hearing loss (CHD7) (26). The doctor may be translated IHH-associated gene studies into novel treatments (31).
Conclusions
The study applied WES and PCR techniques to the screening diagnosis of pathogenic genes of IHH, which provided an indispensable basis for the follow-up clinical treatment of IHH patients and genetic counseling during assisted reproduction. We will further increase the samples and collect the peripheral blood of members to facilitate the discovery of more disease-causing genes.
Acknowledgments
We are extremely grateful to other laboratory members for their valuable comments. Some of the patients in our study were provided by Professor Xiaoyu Yang from Jiangsu Provincial People’s Hospital and Professor Tonghang Guo from the First Affiliated Hospital of the University of Science and Technology of China. We thank the two professors for providing information about IHH patients.
Funding: This work was supported by the Scientific Research Project of Jiangsu Commission of Health (No. H2019065), Key Foundation of Wuhan Huoshenshan Hospital [No. 2020(18)], Clinical Diagnosis and Treatment New Technology Project of Jinling Hospital (No. 22LCZLXJS40), and the Medical Innovation Project of Logistics Service (No. 18JS005).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://tau.amegroups.com/article/view/10.21037/tau-23-225/rc
Data Sharing Statement: Available at https://tau.amegroups.com/article/view/10.21037/tau-23-225/dss
Peer Review File: Available at https://tau.amegroups.com/article/view/10.21037/tau-23-225/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tau.amegroups.com/article/view/10.21037/tau-23-225/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by ethics board of the Jinling Hospital, Nanjing University School of Medicine (No. 2018NZGKJ-003) and informed consent was taken from all the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bianco SD, Kaiser UB. The genetic and molecular basis of idiopathic hypogonadotropic hypogonadism. Nat Rev Endocrinol 2009;5:569-76. [Crossref] [PubMed]
- Palmert MR, Dunkel L. Clinical practice. Delayed puberty. N Engl J Med 2012;366:443-53. [Crossref] [PubMed]
- Grumbach MM. A window of opportunity: the diagnosis of gonadotropin deficiency in the male infant. J Clin Endocrinol Metab 2005;90:3122-7. [Crossref] [PubMed]
- Ali L, Adeel A. Role of basal and provocative serum prolactin in differentiating idiopathic hypogonadotropic hypogonadism and constitutional delayed puberty--a diagnostic dilemma. J Ayub Med Coll Abbottabad 2012;24:73-6. [PubMed]
- Cariboni A, Pimpinelli F, Colamarino S, et al. The product of X-linked Kallmann's syndrome gene (KAL1) affects the migratory activity of gonadotropin-releasing hormone (GnRH)-producing neurons. Hum Mol Genet 2004;13:2781-91. [Crossref] [PubMed]
- Maione L, Dwyer AA, Francou B, et al. GENETICS IN ENDOCRINOLOGY: Genetic counseling for congenital hypogonadotropic hypogonadism and Kallmann syndrome: new challenges in the era of oligogenism and next-generation sequencing. Eur J Endocrinol 2018;178:R55-80. [Crossref] [PubMed]
- Topaloglu AK, Kotan LD. Genetics of Hypogonadotropic Hypogonadism. Endocr Dev 2016;29:36-49. [Crossref] [PubMed]
- Miraoui H, Dwyer AA, Sykiotis GP, et al. Mutations in FGF17, IL17RD, DUSP6, SPRY4, and FLRT3 are identified in individuals with congenital hypogonadotropic hypogonadism. Am J Hum Genet 2013;92:725-43. [Crossref] [PubMed]
- Lima Amato LG, Latronico AC, Gontijo Silveira LF. Molecular and Genetic Aspects of Congenital Isolated Hypogonadotropic Hypogonadism. Endocrinol Metab Clin North Am 2017;46:283-303. [Crossref] [PubMed]
- Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 2010;38:e164. [Crossref] [PubMed]
- Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc 2009;4:1073-81. [Crossref] [PubMed]
- Schwarz JM, Cooper DN, Schuelke M, et al. MutationTaster2: mutation prediction for the deep-sequencing age. Nat Methods 2014;11:361-2. [Crossref] [PubMed]
- Cariboni A, André V, Chauvet S, et al. Dysfunctional SEMA3E signaling underlies gonadotropin-releasing hormone neuron deficiency in Kallmann syndrome. J Clin Invest 2015;125:2413-28. [Crossref] [PubMed]
- Cariboni A, Davidson K, Rakic S, et al. Defective gonadotropin-releasing hormone neuron migration in mice lacking SEMA3A signalling through NRP1 and NRP2: implications for the aetiology of hypogonadotropic hypogonadism. Hum Mol Genet 2011;20:336-44. [Crossref] [PubMed]
- Xu C, Cassatella D, van der Sloot AM, et al. Evaluating CHARGE syndrome in congenital hypogonadotropic hypogonadism patients harboring CHD7 variants. Genet Med 2018;20:872-81. [Crossref] [PubMed]
- Falardeau J, Chung WC, Beenken A, et al. Decreased FGF8 signaling causes deficiency of gonadotropin-releasing hormone in humans and mice. J Clin Invest 2008;118:2822-31. [Crossref] [PubMed]
- Seminara SB, Messager S, Chatzidaki EE, et al. The GPR54 gene as a regulator of puberty. N Engl J Med 2003;349:1614-27. [Crossref] [PubMed]
- Correa-Silva SR, Fausto JDS, Kizys MML, et al. A novel GNRHR gene mutation causing congenital hypogonadotrophic hypogonadism in a Brazilian kindred. J Neuroendocrinol 2018;30:e12658. [Crossref] [PubMed]
- Pankov YA. Kisspeptin and leptin in the regulation of fertility. Mol Biol (Mosk) 2015;49:707-15. [PubMed]
- Kelsh RN. Sorting out Sox10 functions in neural crest development. Bioessays 2006;28:788-98. [Crossref] [PubMed]
- Dai W, Li JD, Zhao Y, et al. Functional analysis of SEMA3A variants identified in Chinese patients with isolated hypogonadotropic hypogonadism. Clin Genet 2020;97:696-703. [Crossref] [PubMed]
- Pingault V, Ente D, Dastot-Le Moal F, et al. Review and update of mutations causing Waardenburg syndrome. Hum Mutat 2010;31:391-406. [Crossref] [PubMed]
- Boehm U, Bouloux PM, Dattani MT, et al. Expert consensus document: European Consensus Statement on congenital hypogonadotropic hypogonadism--pathogenesis, diagnosis and treatment. Nat Rev Endocrinol 2015;11:547-64. [Crossref] [PubMed]
- Nie M, Xu H, Chen R, et al. Analysis of genetic and clinical characteristics of a Chinese Kallmann syndrome cohort with ANOS1 mutations. Eur J Endocrinol 2017;177:389-98. [Crossref] [PubMed]
- Jiang X, Li D, Gao Y, et al. A novel splice site variant in ANOS1 gene leads to Kallmann syndrome in three siblings. Gene 2020;726:144177. [Crossref] [PubMed]
- Topaloğlu AK. Update on the Genetics of Idiopathic Hypogonadotropic Hypogonadism. J Clin Res Pediatr Endocrinol 2017;9:113-22. [PubMed]
- Vissers LE, van Ravenswaaij CM, Admiraal R, et al. Mutations in a new member of the chromodomain gene family cause CHARGE syndrome. Nat Genet 2004;36:955-7. [Crossref] [PubMed]
- Li L, Wang R, Yu Y, et al. CHD7 missense variants and clinical characteristics of Chinese males with infertility. Mol Genet Genomic Med 2020;8:e1372. [Crossref] [PubMed]
- Matsumoto S, Yamazaki C, Masumoto KH, et al. Abnormal development of the olfactory bulb and reproductive system in mice lacking prokineticin receptor PKR2. Proc Natl Acad Sci U S A 2006;103:4140-5. [Crossref] [PubMed]
- Stamou MI, Varnavas P, Plummer L, et al. Next-generation sequencing refines the genetic architecture of Greek GnRH-deficient patients. Endocr Connect 2019;8:468-80. [Crossref] [PubMed]
- George JT, Kakkar R, Marshall J, et al. Neurokinin B Receptor Antagonism in Women With Polycystic Ovary Syndrome: A Randomized, Placebo-Controlled Trial. J Clin Endocrinol Metab 2016;101:4313-21. [Crossref] [PubMed]


