
Integrative analysis of deoxyribonuclease 1-like 3 as a potential biomarker in renal cell carcinoma
Highlight box
Key findings
• DNASE1L3 may be a promising potential biomarker for the diagnosis and treatment of ccRCC patients.
What is known and what is new?
• DNASE1L3 functions as an inhibitor during ccRCC progression, and its expression is significantly correlated with the immune microenvironment of ccRCC.
• The underlying mechanism of this role may be mediated through dephosphorylation of AKT, which suppresses PI3K/Akt pathway activation, impairing ccRCC cell proliferation and invasion.
What is the implication, and what should change now?
• It is uncertain how DNASE1L3 affects the PI3K/Akt pathway, and this mechanism needs further study in the future.
Introduction
Renal cell carcinoma (RCC) is among the top three most common genitourinary malignancies with a worldwide incidence of greater than 400,000 (1,2). Clear cell renal cell carcinoma (ccRCC) is the most common pathological subtype of RCC and accounts for more than 75% of all RCC cases (3). The treatment of RCC relies heavily on surgical resection because RCC is insensitive to radiotherapy and chemotherapy. However, approximately 30% of patients eventually experience recurrence or develop metastases after surgery (4). In recent years, new cancer immunotherapies based on immune checkpoint blockade have been shown to result in survival benefits when used for the treatment of metastatic RCC, thus providing new therapeutic opportunities for patients.
The phosphatidylinositol 3 kinase (PI3K)/protein kinase B (AKT) pathway is a highly conserved and important pathway involving intracellular signal transduction and cellular processes, such as cell proliferation, migration, senescence and apoptosis (5). It has attracted great interest mainly in numerous basic and clinical oncology research efforts since 1987. Usually, extracellular signal stimuli such as growth factors and cytokines activate PI3K and bind to receptor tyrosine kinases (RTKs), leading to phosphorylation of plasma membrane phosphatidylinositol-4,5-bisphosphate (PIP2) to induce phosphatidylinositol-3,4,5-trisphosphate (PIP3). Via its 3’-position, activated PIP3 binds to AKT, and AKT is thus recruited to the plasma membrane to phosphorylate its substrates, activating downstream effector proteins (6). It was reported that the overall genetic alteration rate is 27%, resulting in dysfunction of chromatin remodeling leading to the progression of RCC (7). A series of studies demonstrated that the levels of PI3K/AKT pathway-related proteins were significantly increased and that suppression of this pathway inhibited the proliferation and migration of RCC cell lines (8-10). Presently, a plethora of studies has been dedicated to exploring potential prognostic biomarkers for ccRCC. Notably, some investigations have revealed that ccRCC patients exhibiting low expression of ALDOB (11), high expression of p-CREB1 (12), or elevated expression of SPOCK1 (13) are associated with a less favorable prognosis. However, the association between deoxyribonuclease 1 like 3 (DNASE1L3) and ccRCC remains unexplored.
DNASE1L3, a member of the DNA-degrading enzyme family, was originally discovered to be an endonuclease that cleaves membrane-encapsulated single- and double-stranded DNA during cellular metabolic, necrotic and apoptotic processes (14). Normally, serum DNASE1L3 is involved in maintaining circulating plasma DNA homeostasis by controlling the clearance of fragmentation and end-motif frequencies (15). One study has shown that genetic alterations of DNASLE1L3 resulting in lower endonuclease activity are linked to systemic rheumatic diseases (16). The absence of DNASLE1L3 in serum results in a series of autoimmune inflammatory responses and the production of anti-DNA antibodies, leading to loss of self-tolerance and contributing to the pathogenesis of asthma exacerbations, systemic lupus erythematosus and ankylosing spondylitis (17-19). Accumulating evidence has revealed that DNASE1L3 also plays an important role in tumorigenesis (20-22). Our previous study showed that DNASE1L3 acts as a suppressor of cell cycle progression, proliferation, apoptosis, and metabolism by reducing the activity of rate-limiting enzymes of glycolysis in hepatocellular carcinoma (23). However, the mechanism of DNASEL1L3 in RCC remains unclear and needs to be elucidated.
In the present study, we investigated the correlations between the expression of DNASE1L3 and clinical characteristics, the tumor immune microenvironment and drug sensitivity in ccRCC. Overexpression of DNASE1L3 impaired the proliferation and invasion of ccRCC cells, and this effect may be ascribed to inactivation of the PI3K/AKT pathway. This study indicates that DNASE1L3 may be a potential marker for the diagnosis and target for the treatment of ccRCC. We present this article in accordance with the MDAR and TRIPOD reporting checklists (available at https://tau.amegroups.com/article/view/10.21037/tau-23-355/rc).
Methods
Data preprocessing and patient selection
RNA sequencing (RNA-seq) data of RCC were downloaded from The Cancer Genome Atlas (TCGA) database, and validation microarrays, including GSE36895, GSE53757, and GSE68417, were obtained from the Gene Expression Omnibus (GEO) database. RNA-seq data from TCGA were normalized and analyzed using Edge, and the microarray data in TCGA were analyzed by the Limma R package. The present study included differentially expressed genes (DEGs) that showed a |log2-fold change| >2 and a P value <0.05. Clinical information of RCC patients was downloaded from the TCGA database. The diagnoses of 472 patients with complete clinical baselines were histologically confirmed. Patients with incomplete clinical characteristics, such as clinical stage, histologic grade, pathologic stage or tumor weight, were excluded. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Cell lines and transduction
The normal human renal cell line HK2 and human RCC cell lines ACHN, Caki-1, 786-O, and 769P were purchased from the China Centre for Type Culture Collection (Wuhan, China). All cell lines were cultured following to the guidelines. The lentivirus containing DNASE1L3 overexpressing and the respective lentiviral vector were acquired from GeneCreate Company (Wuhan, China) and transduced into 786-O and ACHN cells using Lipofectamine 3000 (Thermo Fisher Scientific, Waltham, MA, USA) as previously described (23). The transduction efficiency of cells was determined with western blotting following the manufacturer’s instructions.
Cell proliferation assay
786-O and ACHN cells transduced with the DNASE1L3 or empty vector were seeded into 96-well plates and incubated with CCK8 solution following the manufacturer’s protocols. First, 100 µL of cell suspension was added to a 96-well plate. The culture plate was preincubated in an incubator for 24 h (at 37 ℃, 5% CO2); then, 10 µL of renal cancer cell line suspensions were added to the culture plate, and the plate was incubated in the incubator for 0, 24, 48 and 72 h. In each well, 10 µL of CCK-8 solution (Beyotime, China) was introduced, and the culture plate was then placed in an incubator for a duration of 2 hours. Subsequently, the absorbance at 450 nm was determined using a microplate reader.
Cell migration and invasion assays
Cell migration was measured using a wound healing assay. 786-O and ACHN cells transduced with the DNASE1L3 lentiviral vector or empty vector were seeded into 6-well plates in serum-free medium. A 200-µL plastic pipette tip was used to create a scratch after the cells were 80% confluent. The migration capacity of cells was assessed 24 h later under a microscope. The cell invasion ability was tested using a Matrigel chamber (BD Biosciences, USA) following the manufacturer’s protocols. Briefly, cells were seeded into the upper chamber in serum-free medium, and 10% fetal bovine serum was added to the lower chamber to induce cell migration. After 24 h, the cells in the upper chamber were stained with crystal violet and photographed under a microscope.
Western blot analysis
Western blotting was used as previously described (23). Briefly, cellular protein lysate was obtained from the samples and quantified using a BCA Protein Assay Kit (Thermo Fisher Scientific, USA). Then, cellular proteins were separated using SDS–PAGE and transferred to polyvinylidene fluoride membranes. After incubation in 5% nonfat dry milk for 1 h, the membranes were incubated with an anti-DNASE1L3 antibody (Thermo Fisher Scientific, USA, 1:1,000) overnight at 4 ℃. After washing with 20× TBS-tween 20 detergent reagent (Servicebio, China), the membranes were exposed with anti-rabbit/mouse secondary antibodies conjugated with horseradish peroxidase at a dilution of 1:5,000 (Cell Signaling Technology, USA) and captured using an ECL protein detection system (BIO-RAD, USA).
Immunofluorescence assays
Immunofluorescence assays were performed as previously described (23). Briefly, cells were fixed with 4% formaldehyde and permeabilized with 0.1% Triton X-100. Then, the cells were blocked with goat serum and incubated with an anti-P-AKT antibody (Cell Signaling Technology, USA, 1:100) overnight at 4 ℃, followed by an Alexa Fluor 555 (Cell Signaling Technology, USA, 1:200) secondary antibody. The nuclei in the 786-O and ACHN cells were stained with DAPI in the dark, and images were acquired using a fluorescence microscope (Olympus, Japan).
Tumor immune infiltration analysis
Patients were divided into two groups based on the median DNASE1L3 mRNA expression level in the RNA-seq data. The immune infiltration signatures in the high- and low-DNASE1L3 groups were evaluated and plotted using the Tumor IMmune Estimation Resource (TIMER 2.0) and CIBERSORT databases and the single-sample Gene Set Enrichment Analysis (ssGSEA) and Gene Set Variation Analysis (GSVA) R packages (24-27). A Bayes procedure in the EdgeR package was used to identify the DEGs. The immune-related gene list was downloaded from the InnateDB database (28) and intersected with the DEG list. Then, the correlations among DNASE1L3 and the immune-related DEGs and pathways were further explored.
Estimation of the response to drug therapy
The therapeutic target list for RCC was downloaded from a genomics database (29). The correlations between DNASE1L3 expression and drug therapeutic responses were further investigated and plotted with the oncoPredict R package (30).
Statistical analysis
GraphPad Prism software (CA, USA) was used to evaluate the significance of differences between two groups using Student’s t-test and among multiple groups using one-way ANOVA. Pearson correlation analysis was used to analyze the associations between DNASE1L3 expression and the clinical characteristics of patients, the expression of immune-related DEGs, and the infiltration of immune cells. The impact of DNASE1L3 on survival was analyzed and survival curves were plotted by the Kaplan–Meier method. Univariate and multivariate Cox regression analyses were applied to analyze the relationships between clinical parameters and survival times. Based on the significant independent prognostic factors, a nomogram was established with the rms R package to predict the survival probabilities of RCC patients. Calibration curves of 1-, 2- and 3-year survival probabilities and ROC curves were generated to determine the predictive accuracy of the nomogram. The log-rank test was performed to determine statistical significance using R software version 4.2.0. Data in the present study are presented as the mean ± standard error values and were obtained from triplicate experiments. P<0.05 was considered to indicate a significant difference.
Results
DNASE1L3 expression is downregulated in ccRCC
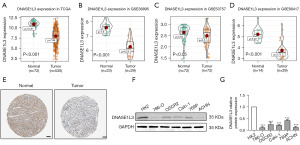
To explore DNASE1L3 expression in ccRCC, the TCGA and GEO databases were used for analysis. As shown in Figure 1A, compared to that in normal samples, DNASE1L3 expression in tumors was significantly lower. Our results of bioinformatics analysis in TCGA were validated using GEO datasets and Human Protein Atlas database (https://www.proteinatlas.org/), revealing that the DNASE1L3 levels as determined by RNA-seq in the normal group were higher than those in the tumor group (Figure 1B-1E). To confirm the findings in these public datasets, normal renal cell and ccRCC cell lines were utilized for western blotting. As indicated in Figure 1F,1G, compared to that in HK2 normal kidney cells, DNASEL13 expression in five kidney tumor cell lines was significantly decreased.
DNASE1L3 expression is correlated with clinical characteristics in ccRCC
To identify the role of DNASE1L3 in ccRCC, we analyzed the correlations among DNASE1L3 expression and clinical characteristics in ccRCC patients. As shown in Figure 2A, DNASE1L3 expression was found to be significantly positively related to patient prognosis. Moreover, our analysis of the clinical characteristics of ccRCC patients indicated that DNASE1L3 expression was significantly related to tumor dimension, tumor weight, clinical stage and histological grade (Figure 2B-2E). The results of Cox proportional hazards regression analyses showed that DNASE1L3 was a significant independent factor in the TGCA cohort (Table S1), indicating that DNASE1L3 might be functionally important in ccRCC pathogenesis.

Construction and validation of the nomogram for ccRCC patient survival
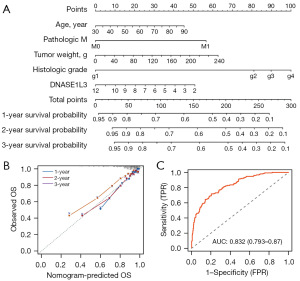
To provide a quantitative method to predict the survival probabilities of ccRCC patients with low and high DNASE1L3 expression, we constructed a nomogram that integrated five independent significant clinicopathological factors (Figure 3A). The calibration curves of 1-, 2- and 3-year survival probabilities and the predictive accuracy of the nomogram (AUC =0.832) indicated that the nomogram was in optimal agreement with an ideal model (Figure 3B,3C).
DNASE1L3 impaired ccRCC cell proliferation and invasion in vitro
Two cell lines were transduced with empty vector and the DNASE1L3-overexpressing lentiviral vector, and the transduction efficiency of DNASEL13 was confirmed by western blotting (Figure S1). To explore the function of DNASE1L3 in ccRCC, ACHN and 786-O cells were used for functional experiments. As indicated in Figure 4A,4B, the CCK8 assay results showed that DNASE1L3 overexpression decreased the proliferative capacity of ACHN and 786-O cells compared to the vector groups. The results showed that overexpression of DNSE1L3 decreased the migration distance of ccRCC cells at 24 h (Figure 4C,4D) compared to that in the vector group. Likewise, the results of the transwell assays were consistent with those of the wound healing assay (Figure 4E,4F). Together, these results demonstrate that DNASE1L3 functions to suppress the proliferation and invasion of ccRCC cells.

DNASE1L3 expression is correlated with immune cell infiltration in ccRCC
Because of the immunoreactive feature of kidney cancer, all samples were classified into two groups based on the median DNASE1L3 expression level, and the associations between DNASE1L3 expression and immune infiltrates were identified. As indicated in Figure 5A and Figure S2, the infiltration of CD8+ T cells, follicular helper T cells, CD4+ T cells, Treg cells, monocytes, macrophages, dendritic cells, mast cells, and neutrophils was related to DNASE1L3 expression. Moreover, the results of TIMER database analysis showed that mutation of DNASE1L3 was significantly linked with infiltration of CD8+ T cells, CD4+ T cells, macrophages and myeloid dendritic cells (Figure S3), and Pearson correlation analysis further indicated that infiltration of CD4+ T cells and macrophages was correlated with DNASE1L3 expression (Figure 5B,5C).

DNASE1L3 expression is linked to immune biomarker expression in ccRCC
To understand the role of DNASEL1L3 in regulating immune activity in ccRCC, immune biomarker data obtained from the InnateDB database were analyzed. As shown in https://cdn.amegroups.cn/static/public/tau-23-355-1.xlsx, the expression of 376 (R>0.3) of the 3,710 immune genes was significantly correlated with DNASEL1L3 expression. The expression levels of the top 20 correlated genes were significantly different between the high- and low-DNASE1L3 groups, as shown in the heatmap (Figure 6A-6C, top right). In addition, the results of GSVA enrichment analysis showed that 19 of 34 pathways were mapped based on the significantly correlated immune genes (Figure 6C and Table S2), and the top 5 signaling networks were the inflammatory response, PI3K/AKT pathway, IL-2/STAT5 pathway, apoptosis and complement pathway.

DNASE1L3 is linked to the PI3K/AKT signaling pathway in ccRCC
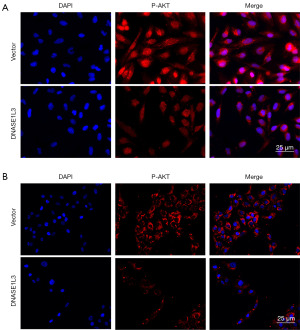
To further determine the potential mechanism of DNASE1L3 in ccRCC, the abundance of the key PI3K/AKT signaling pathway protein P-AKT was tested using immunofluorescence staining. As indicated in Figure 7, compared to the vector group, the abundance of P-AKT was enhanced in the group transduced with DNASE1L3.
DNASE1L3 in connection with drug sensitivity in ccRCC
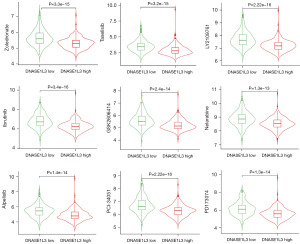
The drug sensitivities of ccRCC samples in the high- and low-DNASE1L3 groups were analyzed based on the DrugBank database. The half-maximal inhibitory concentrations (IC50s) of 143 antitumor drugs were determined, and the nine drugs with the greatest difference in sensitivity were LY2109761, PCI-34051, ibrutinib, taselisib, zoledronate, PD173074, alpelisib, GSK2606414, and nelarabine (Figure 8). Some drugs have been used for the clinical treatment of other tumors; these drugs include ibrutinib, taselisib, zoledronate and alpelisib, and ccRCC samples in the high-DNASE1L3 group exhibited higher sensitivity to these drugs than those in the low-DNASE1L3 group.
Discussion
Because of the heterogeneity among patients, current medical treatments for ccRCC lack suitable efficacy after surgery. In recent decades, numerous methods for antitumor therapy have been developed, but many patients do not obtain durable clinical benefits often due to drug resistance in patients with ccRCC. Therefore, it is essential to further investigate the potential pathogenesis of ccRCC and identify therapeutic targets to guide individualized treatment of patients.
DNASE1L3 is normally expressed in the cytosol, nucleus, endoplasmic reticulum, and extracellular matrix and is involved in balancing DNA cleavage metabolism (23). Many studies have demonstrated the important role of DNASE1L3 mainly in immune-mediated diseases (15,18,22,31). Our previously reported work indicated that DNASE1L3 inhibits tumor progression by promoting apoptosis and inactivating the rate-limiting enzymes of glycolysis in hepatocellular carcinoma (23). Thus, in light of the similarity between these types of solid tumors, it might be of value to explore the function and mechanism of DNASE1L3 in the progression of ccRCC. Thus, we downloaded RNA-seq data from the TCGA and GEO databases and comprehensively analyzed the correlations between DNASE1L3 expression and ccRCC characteristics. The results were consistent with our previous findings that DNASE1L3 expression was significantly decreased in ccRCC samples and that decreased DNASE1L3 expression was significantly positively related to ccRCC prognosis, suggesting the antitumor capacity of DNASE1L3. DNASE1L3 expression has been reported to be associated with tumor grade, tumor size and tumor thrombus formation in liver cancer (31). This result was also found in our present study, which showed that DNASE1L3 expression in ccRCC samples was significantly related to tumor dimension, tumor weight, clinical stage and histological grade. Moreover, the results of functional experiments supported the findings of our bioinformatics analysis, further proving that DNASE1L3 is capable of suppressing cellular proliferation and invasion.
Accumulating studies have proven that the tumor immune microenvironment (TIME) plays a crucial role in regulating oncogenesis. The underlying molecular and cellular features of the TIME affect tumor progression and metastasis by altering the ratios of immunoregulatory components, such as tumor-recruited immune cells and structural stromal components, and shaping an immunosuppressive environment around the tumor to support the evasion of immune surveillance, even that of host immunity (32). One study has revealed that tumor-infiltrating immune cells (TICs), one of the most important components in the tumor environment, are potential biomarkers (33). High infiltration levels of CD8+ T cells and Tregs indicated an unfavorable prognosis for ccRCC (34). Activated CD8+ T and Treg cells interact with antigen-presenting cells, resulting in the production of many cytokines, such as interleukin and vascular endothelial growth factor, to help protect tumor cells against the continuous onslaught of the host immune response (35). This result is consistent with our findings that the infiltration levels of CD8+ T cells and Tregs in the low-DNASE1L3 group were higher than those in the high-DNASE1L3 group, suggesting that high DNASE1L3 expression might impair tumor immune evasion ability. Moreover, the results showed that the infiltration levels of T follicular helper cells, monocytes, macrophages, dendritic cells, mast cells, and neutrophils were different between the high- and low-DNASE1L3 groups, indicating that DNASE1L3 expression might influence the immune response during ccRCC progression.
The PI3K-AKT signaling pathway plays a crucial role in regulating tumor progression and metastasis by influencing cell proliferation, migration and invasion (36). The results derived from preclinical models and clinical trials of mice also indicated that suppression of the PI3K-AKT signaling pathway impaired tumor immunosurveillance through inactivation of immunosuppressive pathways and promotion of antitumor immune responses (37). The findings from breast cancer studies showed that inactivating the PI3K-AKT signaling network with a PI3K inhibitor weakened the interaction of PD-L1 with PD-1, which enhanced the proliferation and migration capacities of T cells (38). Consistent with previous findings, the results above showed that the PI3K-AKT signaling pathway was mapped by both gene set analyses based on the significantly correlated immune genes between the high- and low-DNAE1L3 groups. The results of western blot analysis revealed that overexpressing DNAE1L3 reduced the P-AKT level, suggesting that DNASE1L3 expression was correlated with the tumor immune microenvironment possibly via regulation of the PI3K/Akt pathway in RCC. In addition, based on the successful curative effects of immune checkpoint inhibitors in cancers such as melanoma and leukemia, a series of innovative drugs are currently being used in the clinical treatment of RCC patients. In this regard, we found that the IC50 values of ibrutinib, taselisib, zoledronate and alpelisib were extremely low in samples with high expression of DNASEL13. However, this research has several limitations, not least the accuracy of DNASE1L3 prediction was not validated by applying clinical cases of RCC. Furthermore, comprehensive animal experiments and in-depth studies are yet to be conducted, which would elucidate the specific mechanisms underlying the cancer-inhibitory effects of DNASE1L3. Additionally, it is crucial to acknowledge that the database samples utilized in this research may potentially contain outliers, impacting the generalizability of the results.
Conclusions
This study reveals that DNASE1L3 functions as an inhibitor during ccRCC progression and is significantly correlated with the immune microenvironment of ccRCC. The underlying mechanism of this role may involve dephosphorylation of AKT, thus leading to suppression of PI3K/Akt pathway activation and impairment of ccRCC cell proliferation and invasion. However, it is uncertain how DNASE1L3 affects the PI3K/Akt pathway, and we will study this mechanism further in the future.
Acknowledgments
Funding: This work was supported by the National Natural Science Foundation of China (No. 82100703), Natural Science Foundation of Hubei Province (No. 2022CFC015) and the Funds Project for Young Teachers of Wuhan University in Central Universities (Nos. 2042021kf0099 and 2042021kf0155).
Footnote
Reporting Checklist: The authors have completed the MDAR and TRIPOD reporting checklists. Available at https://tau.amegroups.com/article/view/10.21037/tau-23-355/rc
Data Sharing Statement: Available at https://tau.amegroups.com/article/view/10.21037/tau-23-355/dss
Peer Review File: Available at https://tau.amegroups.com/article/view/10.21037/tau-23-355/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tau.amegroups.com/article/view/10.21037/tau-23-355/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Jonasch E, Walker CL, Rathmell WK. Clear cell renal cell carcinoma ontogeny and mechanisms of lethality. Nat Rev Nephrol 2021;17:245-61. [Crossref] [PubMed]
- Castro DV, Malhotra J, Meza L, et al. How to Treat Renal Cell Carcinoma: The Current Treatment Landscape and Cardiovascular Toxicities. JACC CardioOncol 2022;4:271-5. [Crossref] [PubMed]
- Moch H, Amin MB, Berney DM, et al. The 2022 World Health Organization Classification of Tumours of the Urinary System and Male Genital Organs-Part A: Renal, Penile, and Testicular Tumours. Eur Urol 2022;82:458-68. [Crossref] [PubMed]
- Deleuze A, Saout J, Dugay F, et al. Immunotherapy in Renal Cell Carcinoma: The Future Is Now. Int J Mol Sci 2020;21:2532. [Crossref] [PubMed]
- Fruman DA, Chiu H, Hopkins BD, et al. The PI3K Pathway in Human Disease. Cell 2017;170:605-35. [Crossref] [PubMed]
- Liu P, Cheng H, Roberts TM, et al. Targeting the phosphoinositide 3-kinase pathway in cancer. Nat Rev Drug Discov 2009;8:627-44. [Crossref] [PubMed]
- Guo H, German P, Bai S, et al. The PI3K/AKT Pathway and Renal Cell Carcinoma. J Genet Genomics 2015;42:343-53. [Crossref] [PubMed]
- Liu W, Yan B, Yu H, et al. OTUD1 stabilizes PTEN to inhibit the PI3K/AKT and TNF-alpha/NF-kappaB signaling pathways and sensitize ccRCC to TKIs. Int J Biol Sci 2022;18:1401-14. [Crossref] [PubMed]
- Li B, Zhang X, Ren Q, et al. NVP-BEZ235 Inhibits Renal Cell Carcinoma by Targeting TAK1 and PI3K/Akt/mTOR Pathways. Front Pharmacol 2022;12:781623. [Crossref] [PubMed]
- Mao W, Wang K, Xu B, et al. ciRS-7 is a prognostic biomarker and potential gene therapy target for renal cell carcinoma. Mol Cancer 2021;20:142. [Crossref] [PubMed]
- Shao Y, Wu B, Yang Z, et al. ALDOB represents a potential prognostic biomarker for patients with clear cell renal cell carcinoma. Transl Androl Urol 2023;12:549-71. [Crossref] [PubMed]
- Zhang Z, Guan B, Li Y, et al. Increased phosphorylated CREB1 protein correlates with poor prognosis in clear cell renal cell carcinoma. Transl Androl Urol 2021;10:3348-57. [Crossref] [PubMed]
- Chen J, Ye Z, Liu L, et al. Assessment of the prognostic value of SPOCK1 in clear cell renal cell carcinoma: a bioinformatics analysis. Transl Androl Urol 2022;11:509-18. [Crossref] [PubMed]
- Inokuchi S, Mitoma H, Kawano S, et al. Homeostatic Milieu Induces Production of Deoxyribonuclease 1-like 3 from Myeloid Cells. J Immunol 2020;204:2088-97. [Crossref] [PubMed]
- Serpas L, Chan RWY, Jiang P, et al. Dnase1l3 deletion causes aberrations in length and end-motif frequencies in plasma DNA. Proc Natl Acad Sci U S A 2019;116:641-9. [Crossref] [PubMed]
- Herrera-Luis E, Lorenzo-Diaz F, Samedy-Bates LA, et al. A deoxyribonuclease 1-like 3 genetic variant associates with asthma exacerbations. J Allergy Clin Immunol 2021;147:1095-1097.e10. [Crossref] [PubMed]
- Coke LN, Wen H, Comeau M, et al. Arg206Cys substitution in DNASE1L3 causes a defect in DNASE1L3 protein secretion that confers risk of systemic lupus erythematosus. Ann Rheum Dis 2021;80:782-7. [Crossref] [PubMed]
- Hartl J, Serpas L, Wang Y, et al. Autoantibody-mediated impairment of DNASE1L3 activity in sporadic systemic lupus erythematosus. J Exp Med 2021;218:e20201138. [Crossref] [PubMed]
- Sun Y, Ouyang B, Xie Q, et al. Serum Deoxyribonuclease 1-like 3 is a potential biomarker for diagnosis of ankylosing spondylitis. Clin Chim Acta 2020;503:197-202. [Crossref] [PubMed]
- Wang S, Ma H, Li X, et al. DNASE1L3 as an indicator of favorable survival in hepatocellular carcinoma patients following resection. Aging (Albany NY) 2020;12:1171-85. [Crossref] [PubMed]
- Chen J, Ding J, Huang W, et al. DNASE1L3 as a Novel Diagnostic and Prognostic Biomarker for Lung Adenocarcinoma Based on Data Mining. Front Genet 2021;12:699242. [Crossref] [PubMed]
- Deng Z, Xiao M, Du D, et al. DNASE1L3 as a Prognostic Biomarker Associated with Immune Cell Infiltration in Cancer. Onco Targets Ther 2021;14:2003-17. [Crossref] [PubMed]
- Xiao Y, Yang K, Liu P, et al. Deoxyribonuclease 1-like 3 Inhibits Hepatocellular Carcinoma Progression by Inducing Apoptosis and Reprogramming Glucose Metabolism. Int J Biol Sci 2022;18:82-95. [Crossref] [PubMed]
- Li T, Fu J, Zeng Z, et al. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res 2020;48:W509-14. [Crossref] [PubMed]
- Chen B, Khodadoust MS, Liu CL, et al. Profiling Tumor Infiltrating Immune Cells with CIBERSORT. Methods Mol Biol 2018;1711:243-59. [Crossref] [PubMed]
- Yi M, Nissley DV, McCormick F, et al. ssGSEA score-based Ras dependency indexes derived from gene expression data reveal potential Ras addiction mechanisms with possible clinical implications. Sci Rep 2020;10:10258. [Crossref] [PubMed]
- Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics 2013;14:7. [Crossref] [PubMed]
- Breuer K, Foroushani AK, Laird MR, et al. InnateDB: systems biology of innate immunity and beyond--recent updates and continuing curation. Nucleic Acids Res 2013;41:D1228-33. [Crossref] [PubMed]
- Yang W, Soares J, Greninger P, et al. Genomics of Drug Sensitivity in Cancer (GDSC): a resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res 2013;41:D955-61. [Crossref] [PubMed]
- Maeser D, Gruener RF, Huang RS. oncoPredict: an R package for predicting in vivo or cancer patient drug response and biomarkers from cell line screening data. Brief Bioinform 2021;22:bbab260. [Crossref] [PubMed]
- Chen QY, Li L, Suo DQ. DNase1L3 suppresses hepatocellular carcinoma growth via inhibiting complement autocrine effect. Neoplasma 2021;68:683-91. [Crossref] [PubMed]
- Galland S, Stamenkovic I. Mesenchymal stromal cells in cancer: a review of their immunomodulatory functions and dual effects on tumor progression. J Pathol 2020;250:555-72. [Crossref] [PubMed]
- Wang J, Li Z, Gao A, et al. The prognostic landscape of tumor-infiltrating immune cells in cervical cancer. Biomed Pharmacother 2019;120:109444. [Crossref] [PubMed]
- Giraldo NA, Becht E, Vano Y, et al. Tumor-Infiltrating and Peripheral Blood T-cell Immunophenotypes Predict Early Relapse in Localized Clear Cell Renal Cell Carcinoma. Clin Cancer Res 2017;23:4416-28. [Crossref] [PubMed]
- Hoyer S, Prommersberger S, Pfeiffer IA, et al. Concurrent interaction of DCs with CD4(+) and CD8(+) T cells improves secondary CTL expansion: It takes three to tango. Eur J Immunol 2014;44:3543-59. [Crossref] [PubMed]
- O'Donnell JS, Massi D, Teng MWL, et al. PI3K-AKT-mTOR inhibition in cancer immunotherapy, redux. Semin Cancer Biol 2018;48:91-103. [Crossref] [PubMed]
- Xue G, Zippelius A, Wicki A, et al. Integrated Akt/PKB signaling in immunomodulation and its potential role in cancer immunotherapy. J Natl Cancer Inst 2015;107:djv171. [Crossref] [PubMed]
- Mittendorf EA, Philips AV, Meric-Bernstam F, et al. PD-L1 expression in triple-negative breast cancer. Cancer Immunol Res 2014;2:361-70. [Crossref] [PubMed]


