Proteomic approach towards identification of seminal fluid biomarkers from individuals with severe oligozoospermia, cryptozoospermia and non-obstructive azoospermia: a pilot study
Highlight box
Key findings
• The presented study has indicated ADGRG2, SLC2A3 and SMS protein as potential seminal plasma biomarkers of diminished spermatogenesis.
What is known and what is new?
• Seminal plasma proteins may reflect the condition of male reproductive health, including spermatogenesis status.
• Here, we report on the seminal plasma ADGRG2 protein as an attractive biomarker for severe oligozoospermia, cryptozoospermia and azoospermia and on SLC2A3 and SMS proteins as potential biomarkers of non-obstructive azoospermia however, with different molecular background.
What is the implication, and what should change now?
• The reported proteins may be included in potential diagnostic panels dedicated for the assessment of reasons of failing spermatogenesis however, they may not fully explain mechanisms of infertility. The potential diagnostic panel should be enriched in other biomarkers enabling correct diagnosis and application of targeted therapy.
Introduction
In recent years, male infertility has become a real medical and sociological problem of a modern world. It is mainly due to deteriorating semen parameters like reduced number of spermatozoa, their motility or morphology. Among these, total lack of spermatozoa in the semen is the most difficult one for successful therapy. The reduced number or absence of spermatozoa in an ejaculate can be a result of either anatomical blockage within male reproductive tract (obstruction at the level of vas deference, epididymis or ejaculatory ducts) (1) or spermatogenesis disruption. The later one may occur due to genetic mutations, hormonal imbalance, incomplete testis development or varicocele. These factors may lead to the total lack of spermatozoa in the testes or arrested spermatogenesis (2). Nevertheless, not all the factors of reduced spermatogenesis have been identified yet.
Seminal plasma is a mixture of secretions released during activity of the testis and accessory sex glands, within male reproductive tract. The fluid comprises 80–90% of the ejaculate and its role is to support spermatozoa with an environment enabling final fertilization success (3). As seminal plasma is abundant in proteins, a number of studies aimed to find reliable biomarkers reflecting the conditions of male reproductive tract and sperm quality in different animal species (4-6), including humans (7-9). The studies on seminal biomarkers concerned stallion sperm motility (10), human spermatogenesis (11) and/or azoospermia (12). The proteomic approach was also applied regarding the prediction of in vitro fertilization (IVF) success (13). Among these all, the most attractive option is the possible use of seminal fluid as convenient for handling of biological material containing biomarkers being helpful for diagnosis of spermatogenesis and possible therapy monitoring.
Non-obstructive azoospermia (NOA), cryptozoospermia (C) and severe oligozoospermia (SO) are diseases very often linked to genomic aberrations (14-17). However, such diagnostic attempt usually requires invasive testicular open biopsy which is rather a controversial issue. Thus, seminal plasma proteins are still of high interest for clinicians and infertility researchers. The optimal seminal fluid biomarker should distinguish among different types of male infertility and/or indicate their etiology. Despite of many studies on infertility biomarkers, still the successful protein candidates have not been selected. Taking into account sperm cells, proAKAP4 has become a commercial biomarker of sperm quality predicting successful fertilization (18-20). Unfortunately, yet no seminal fluid protein has become an universal and/or commercial marker for male infertility. One of the prospective seminal plasma biomarkers is Heat shock-related 70 kDa protein 2 (HSPA2) which is a testis-enriched molecule involved in spermatogenesis (11). According to our previous data, HSPA2 is a biomarker of azoospermia and in some cases of C (11). Yao et al. (12) indicated seminal fluid sodium-coupled monocarboxylate transporter 2 (SLC5A12) as a protein diversifying NOA and obstructive azoospermia (OA) while Histone H2B type 1-A (H2BC1) as a predictive factor for efficient sperm retrieval from NOA patients. Légaré et al. (21) found cysteine-rich secretory protein 1 (CRISP1) as a marker of OA whereas Davalieva et al. (8) described fibronectin (FN1), prostatic acid phosphatase (PAP), proteasome subunit alpha type-3 (PSMA3), beta-2-microglobulin (B2M), galectin-3-binding protein (LGALS3BP), prolactin-inducible protein (PIP) and cytosolic nonspecific dipeptidase (CNDP2) as protein biomarkers of azoospermia.
Different approaches can be used for identification of protein markers, ranging from biochemical assays (22) to high throughput proteomics (23). The biochemical (or immunological) assays are usually limited to a small number of proteins whereas proteomics ensures the analysis of large number of proteins simultaneously however, it can also generate false positive results (24). In our study, we have used nanoUPLC-MS/MS followed by a robust peptide quantification analysis (25,26) and an extensive Western blotting (WB) validation to examine proteins from seminal fluid of patients with NOA, C, and SO. We present this article in accordance with the MDAR reporting checklist (available at https://tau.amegroups.com/article/view/10.21037/tau-23-130/rc).
Methods
Reagents
The reagents were purchased from Sigma-Aldrich ChemieGmbh (Munich, Germany), unless stated otherwise.
Seminal plasma
The samples of seminal plasma were received from healthy, normozoospermic volunteers and from males having problems with fertility. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the National Bioethical Committee, Ministry of Health, Warsaw, Poland; code: OkB-5-2/15 (No. OKB.078.6.2015) and informed consent was obtained from all individual participants. The samples were collected by masturbation following 3–5 days of sexual abstinence. The routine seminal analysis was performed according to World Health Organization (27) guidelines. The samples were classified into four groups under study: the control group (normozoospermic samples), patients with SO, C and NOA. SO and C were also non-obstructive cases. Seminal plasma was prepared for analysis as already described (11).
Mass spectrometry protein identification
Seminal plasma samples from normozoospermic males (n=10) and from patients with SO (n=10), C (n=7) and NOA (n=17) were subjected to comparative proteomic analysis. Protein identification and quantification were performed in Mass Spectrometry Lab at the Institute of Biochemistry and Biophysics (IBB), Polish Academy of Sciences.
Sample preparation
After being reduced with 0.5 M (5 mM f.c.) Tris(2-carboxyethyl)phosphine hydrochloride (TCEP; 1 h, 60 ℃) and blocked with 200 mM S-methyl methanethiosulfonate (MMTS, 10 mM f.c.; 10 min, RT), the protein mixtures were trypsinized overnight with 10 µL of 0.1 µg/uL trypsin. Peptide mixtures were separated with RP‑18 pre-column (Waters, Milford, MA, USA) using 0.1% formic acid (FA) and then transferred to a nano-HPLC RP-18 column (internal diameter 75 µM, Waters, Milford, MA, USA) using a gradient of solvent B (0–35% can, 160 min) with solvent A being 0.1 % FA at a flow rate of 250 nL/min. The column setup was coupled to the Q Exactive mass spectrometer (Thermo Electron Corp., San Jose, CA, USA). For quantification, the samples were evaluated twice, once in data-dependent acquisition mode and once as a profile of LC-MS spectrum.
Analysis of mass spectrometry data
The recorded fragmentation spectra were analyzed with Mascot Distiller software (v. 2.6, MatrixScience, London, UK) and searched with the Mascot Search Engine (MatrixScience, London, UK, Mascot Server 2.5) against the human proteins deposited in Uniprot database (version 20170927, 71,579 sequences; 24,126,051 residues). The peptide and fragment mass tolerance settings were established as already described (26). The following research parameters were applied: enzyme: Trypsin, missed cleavages: 1; fixed modifications: Methylthio (C); variable modifications: Oxidation (M); instrument: higher-energy collisional dissociation (HCD). The target/decoy database search approach was applied for the peptide assignments (28) followed by the mass calibration and data filtering using MScan software—a developed in-house software (http://proteom.ibb.waw.pl/mscan/) already described elsewhere (29).
Quantification
The quantification was performed in the same way as already described by Banaś et al. (30) using an in-house quantification platform of IBB (25) and Diffprot software (26) with the following settings: number of random peptide sets: 106; clustering of peptide sets: only when 90% identical; normalization: LOWESS.
Bioinformatic analysis
An enriched functional analysis of proteomes for each tested group of samples was done using David software and the protein products were annotated to the following Gene Onthology (GO) terms: biological process, cellular compartmentalization and molecular function.
Western blot validation
The validation of MS results was performed for normozoospermic controls (n=10), patients with SO (n=10), C (n=6) and NOA (n=13). The preparation of seminal plasma samples and immunoblotting were performed as in our previous studies (11,31). The following primary antibodies were selected for immunodetection: anti-ADGRG2 (1:600, HPA001478), anti-SEMG2 (Abcam, Cambridge, Great Britain; 1:500, ab108085), anti-SLC2A3 (Abcam; 1:500, ab191071), anti-spermine synthase (SMS) (Abcam; 1:1,000, ab156879), anti-Ras-related protein Rab-3B (RAB3B) (Abcam; 1:1,000, ab177949) and anti-lactotransferrin (LTF) (Millipore; 1:2,000, 07-685). The secondary antibody, horseradish peroxidase-conjugated Goat anti-rabbit IgG H&L (ab97051, Abcam) was added: in 1:40,000 (for anti-LTF, anti-SEMG2) or 1:10,000 (for anti-RAB3B, anti-SLC2A3 and anti-ADGRG2) dilutions. A representative (most often K11) control seminal plasma sample was set as an internal reference in the respective tested groups for Western blot analyses. The images were captured with the ChemiDoc™ MP System (Bio-Rad Laboratories) and were analyzed with Image Lab 6.0.1 software (Bio-Rad Laboratories) applying total protein normalization.
Statistical analysis
Statistical analysis of WB signals was performed with GraphPad Prism 7 software. The calculations were done as grouped Mann-Whitney analyses.
Results
Mass spectrometry
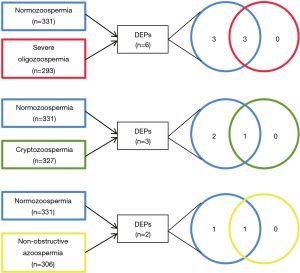
Altogether, the nanoUPLC-MS/MS analysis enabled the identification of 368 different proteins (see https://cdn.amegroups.cn/static/public/tau-23-130-1.xlsx) from seminal plasma. The distribution among the studied groups was as follows: 331 proteins in a group with normozoospermia, 293 in males with SO, 327 in males with C and 306 in males with NOA (Figure 1). The quantitative analysis revealed differentially expressed proteins (DEPs) in seminal plasma from infertile patients and there were identified 6, 3 and 2 DEPs among patients with SO, C and NOA, respectively (Table 1 and Figure 2).

Table 1
| Groups | Protein name | Protein ID | Peptide number | q value | Ratio infertile/controls |
|---|---|---|---|---|---|
| Severe oligozoospermia | Hypoxia up-regulated protein 1 (HYOU1) | A0A087X054 Q9Y4L1 |
2 | NA | Only in control |
| Spermine synthase (SMS) | P52788 | 2 | NA | Only in control | |
| Solute carrier family 2, facilitated glucose transporter member 3 (SLC2A3) | P11169 | 2 | NA | Only in control | |
| Semenogelin-1 (SEMG1) | P04279 | 109 | 0.00029 | 1.72 | |
| Semenogelin-2 (SEMG2) | Q02383 | 151 | 0.00013 | 1.47 | |
| Adhesion G-protein coupled receptor G2 (ADGRG2) | Q8IZP9 | 4 | 0.04313 | 0.51 | |
| Cryptozoospermia | Ras-related protein Rab-3B (RAB3B) | P20337 | 2 | NA | Only in control |
| Plasma alpha-L-fucosidase (FUCA2) | Q9BTY2 | 2 | NA | Only in control | |
| Lactotransferrin (LTF) | E7EQB2 E7ER44 P02788 |
58 | 0.06560 | 1.32 | |
| Non-obstructive azoospermia | Solute carrier family 2, facilitated glucose transporter member 3 (SLC2A3) | P11169 | 2 | NA | Only in control |
| Cadherin-1 (CDH1) | A0A087WX17 A0A087WU43 P12830 A0A087WXI5 |
3 | 0.06188 | 2.08 |
The listed proteins are of q<0.01, DEPs, differentially expressed proteins; NA, not available.
Bioinformatic analysis
The investigated GO terms revealed that the seminal plasma proteomes of the all tested groups were similar regarding their involvement in biological processes, cellular compartmentalization and molecular function (Figure S1).
As there was a limited number of identified DEPs among three groups of patients under study, the enriched functional analysis by means of bioinformatics tools was possible only in the group of males with SO (Table 2).
Table 2
| Category | Term | Genes | Benjamini correction |
|---|---|---|---|
| GOTERM_CC_DIRECT | Extracellular exosome | SMS, HYOU1, SLC2A3, SEMG1, SEMG2, ADGRG2 | 1.6E−3 |
| GOTERM_BP_DIRECT | Coagulation | SEMG1, SEMG2 | 1.2E−2 |
| GOTERM_BP_DIRECT | Positive regulation of serine-type endopeptidase activity | SEMG1, SEMG2 | 1.2E−2 |
| GOTERM_BP_DIRECT | Negative regulation of sperm motility | SEMG1, SEMG2 | 1.3E−2 |
| GOTERM_BP_DIRECT | Antibacterial humoral response | SEMG1, SEMG2 | 8.8E−2 |
| GOTERM_BP_DIRECT | Protein heterooligomerization | SEMG1, SEMG2 | 1.1E−1 |
| UP_KEYWORDS | Signal | HYOU1, SEMG1, SEMG2, ADGRG2 | 9.8E−1 |
| GOTERM_CC_DIRECT | Extracellular region | HYOU1, SEMG1, SEMG2 | 5.9E−1 |
| UP_KEYWORDS | Glycoprotein | HYOU1, SLC2A3, SEMG2, ADGRG2 | 9.8E−1 |
DEPs, differentially expressed proteins; SO, severe oligozoospermia.
Western blot analysis
The results of WB validation differed slightly from quantitative analysis of MS data however, immunoblotting is much more sensitive technique and therefore it provides more precise protein identification. In Figure 3, there are presented WB images of proteins of which differences in their signal intensity were of statistical importance; in Figure 4, there are presented images of proteins of which differences in their signal intensity were out of significant importance.


Discussion
The aim of the present study was to identify seminal plasma proteins having a potential to serve as biomarkers for spermatogenesis disorders. For this purpose, a quantitative proteomic approach combined with immunological assessment was applied.
For the quantitative analysis of proteomic data, the application of statistical Diffprot analysis was applied reducing possible false positive outcomes (26). As a result, in the analysis was selected relatively small number of proteins defined as DEPs among patients under study and these were HYOU, SMS, SLC2A3, SEMG1, SEMG2, and ADGRG2 for patients with SO; RAB3B, FUCA2 and LTF for C patients; and SLC2A3 and CDH1 for NOA patients (Table 1). The functional annotation analysis was possible only for patients with SO due to too little amount of proteins identified in the seminal plasma samples from the remaining infertile patients (C and NOA). According to this analysis, most of DEPs in SO patients was of exosomal origin (Table 2) what is in line with general knowledge of seminal fluid composition (30,31). The immunological assessment was performed with SMS, SLC2A3, ADGRG2, RAB3B and LTF protein products. After WB identification, only two DEPs (ADGRG2 and RAB3) were of statistical significance (Figure 3). The remaining DEPs was out of significance among all the groups of patients analyzed however, there was strong heterogeneity among samples from the NOA patients cohort forcing us to discuss all the DEPs identified.
ADGRG2
Quantitative proteomics identified ADGRG2 as DEP exclusively significant for seminal plasma of SO patients (Table 1) but the immunoblotting indicated this protein to be significantly deregulated also in seminal plasma of C and NOA patients (Figure 3). The Western blot analysis showed that ADGRG2 was expressed heterogeneously among control samples as well as in SO and NOA individuals and there was no identifiable signal for this protein in C patients. ADGRG2 is an extracellular and transmembrane protein involved in G protein-coupled receptor signaling pathway. The protein is widely expressed in human epididymis [it is also known as human epididymis-specific protein 6 (He6)], however, it was also identified in efferent ductuli (32) and other tissues of human body (33-35). Its localization within a cell is apical and in efferent ductuli and proximal epididymis is limited to non-ciliated cells (32,33). Adgrg2 knockout (KO) mice were shown to exhibit simultaneous downregulation of epididymal transcripts for proteins (36) that were later proven to be involved in processes regulating sperm maturation, e.g., lipid raft movement, Ca2+ homeostasis, acquisition of the capability of fertilization (37-39). An advanced set of experiments performed by Zhang et al. (40) established the presence of a multimeric complex governing the regulation of ionic and pH homeostasis in efferent ductuli and proximal epididymis. This complex was created by the interaction between ADGRG2, Gq, beta-arrestin 1 and cystic fibrosis transmembrane conductance regulator (CFTR) ion channel among which beta-arrestin 1 was shown to act as a scaffold for ADGRG2/CFTR complex in the apical membranes of non-ciliated cells. According to the study (40), the coupling between proteins Gq and ADGRG2 enables the regulation of Cl− currents via CFTR as the reduction in either ADGRG2, beta-arrestin1 or Gq protein finally resulting in decreased constitutive CFTR currents and imbalance in pH homeostasis. Thus, ADGRG2 is claimed to be involved in fluid resorption in efferent ductuli and initial segment of epididymis (40). Indeed, a targeted degradation of one of the transmembrane domains of ADGRG2 in male mice resulted in disregulation of fluid resorption and consequently, accumulation of fluid in testes and stasis of spermatozoa within efferent ducts (41). In the literature, there were reported different types of ADGRG2 mutations that are linked to congenital absence of the vas deferens (CAVD) and thus, obstructive azoospermia (42-46). It is estimated that ADGRG2 mutations comprise 2% of all CAVD cases (47) however, they used to accompany mutations in CFTR gene which is assessed to be the main pathogenic gene in CAVD (42). In our study, we showed that ADGRG2 is present also in human seminal plasma (secretory ADGRG2, sADGRG2) (Figure 3) and that all the C patients under study had no ADGRG2 in their seminal plasma samples. It is very probable that C in these patients was a cause of inhibited ADGRG2 expression due to mutation in this gene. It can be assumed that spermatogenesis in the C patients was active but the problems with pH homeostasis and fluid resorption in efferent ductuli and epididymis (place of sperm maturation) resulted in obstructive fluid stasis and drastic reduction in sperm count. To prove this hypothesis there would be required testis and/or epididymis biopsy from all the C patients but, on the other hand, it can be supported by the research by Wang et al. (45) in which CAVD patients had active spermatogenesis (successful MESA or TESE) but the extracted spermatozoa were of low quality. It is very probable that ADGRG2 may also play a role in NOA but due to its heterogeneous content in seminal plasma such statement would require a study including much bigger cohort.
RAB3B
In our MS analysis, RAB3B was found to be not present in semen plasma of C patients (Table 1) however, WB showed this protein to be only downregulated in this group of individuals (Figure 3). Reduced levels of RAB3B were also detected by WB in patients with SO and NOA (Figure 3).
Rab proteins are GTP-binding proteins that are involved in exocytosis and localization of other proteins towards plasma membrane (48). Some of them were found to be involved in female and male meiosis (49). They localized close to Golgi apparatus, plasma membrane and cell junctions. At the protein level, the RAB3B isoform is mainly expressed in gastrointestinal tract, pancreas, prostate and placenta but its high mRNA content was also detected in brain tissues (according to Human Protein Atlas). Other RAB3 isoforms (RAB3A, RAB3C and RAB3D) have been reported to be involved in oncogenesis however, there is no knowledge on the oncogenic role of RAB3B (48). Currently, several reports indicated various signaling pathways and mechanisms engaging RAB3B, i.e., in human epithelial cells Rab effector Noc2 protein (NOC2) was documented to be an effector of RAB3B (50) and in human platelets RAB3B was documented to bind calmodulin in Ca2+-dependent manner (51). Looking at the results from the present study (Table 1, Figure 3) it can be concluded that reduced sperm number in ejaculate may be also associated with disturbances in exocytosis (see Table 2 for SO).
LTF
Label free proteomics indicated LTF to be slightly (1.32 times) upregulated in semen plasma of C patients (Table 1) however, WB analysis showed no significant differences for that protein amount in any group of patients under study (Figure 4). Nonetheless, there was clearly visible lack of the lower LTF band in the WB image for several NOA individuals (Figure 4).
LTF is a glycoprotein expressed in epididymis and prostate gland and secreted to the semen in seminal vesicles (52,53). LTF can attach to the surface of spermatozoa via its receptor (54). Generally, LTF displays antioxidant and antibacterial properties due to its ability to bind iron. Indeed, numerous data reported altered LTF levels in semen samples exposed to oxidative stress conditions (31,55,56). In our previous study on asthenozoospermia, there was reported an increased level of LTF in low-motile spermatozoa accompanied by elevated production of ROS however, there was no significant difference in the levels of semen plasma LTF between control and asthenozoospermic samples (31). Nonetheless, a protective role of LTF to spermatozoa was clearly documented in the literature (57,58). The presence of two bands (~65 and ~78 kDa) for semen plasma LTF is in an accordance to our previous study (31). These bands can represent two LTF isoforms being a result either of different levels of protein glycosylation or the presence of an additional truncated form (~65 kDa) of LTF. It can be assumed that in NOA patients there is a tendency of the lower mass LTF isoform to disappear.
Solute carrier family 2, facilitated glucose transporter member 3 (SLC2A3)
In our study, the MS analysis has indicated the SLC2A3 not to be present in seminal fluid samples from SO an NOA patients (Table 1) but the WB showed this protein to be strongly reduced only in the NOA cohort (Figure 4). The grouped statistical analysis showed this depletion was out of significance however, the WB image clearly shows the presence of SLC2A3 only in 2 out of 13 samples from NOA patients (Figure 4).
According to the Human Protein Atlas, SLC2A3 is mainly expressed in cerebral cortex, lungs, testis, epididymis, placenta, bone marrow and lymphoid tissues. It is also known as Glucose transporter type 3 (GLUT3) and is an integral component of plasma membrane. In a study by Soudmand et al. (57) using a mouse model, animals subjected to a high-intense exercise training protocol had a diminished level of Slc2a3 in their Sertoli cells and increased in their germ cells. Based on immunohistochemical staining, the mice displayed impaired spermato- and spermiogenesis. The authors explained this phenomenon by reduced glucose uptake and inhibited export of lactate within Sertoli cells what resulted in disturbed sperm development. As the lactate is the preferred energy substrate for developing germ cells (58), it seems that diminished SLC2A3 levels in Sertoli cells disturb their nurturing of spermatozoa. Additionally, it seems that the expression of SLC2A3 in testes can be regulated at the epigenetic level. Indeed, the upregulated expression of mmu-miR-320-3p (the murine homolog of hsa-miR-320c, one of the most upregulated miRNAs in testicular biopsies from SC-only syndrome (SCOS) (59) inhibited Slc2a3 expression in murine Sertoli cells and contributed to germ cell loss driven by Sertoli cell dysfunction (58). Nonetheless, not only epigenetics is reported to have an impact on SLC2A3 level. It is very well established that sex steroid hormones, including 5α-dihydrotestosteron, have an impact on the glucose uptake and the expression of glucose transporters in Sertoli cells (60,61). It has been also documented that a deficiency in thyroid hormones in early developmental period also affects this transport due to reduced Slc2A3 expression and finally leads to the apoptosis of male germ cells (62). As it was documented that in human males SLC2A3 level is considerably higher in testes than in epididymis (63), we may conclude that the lack of this protein in 11 (out of 13) seminal plasma NOA samples is connected with dysfunctional testes (probably due to Sertoli cells dysfunction) and may be a reason of azoospermia.
SMS
The quantitative MS approach have indicated SMS to be not present in seminal plasma from SO patients (Table 1) however, the validation by WB showed this protein to be present in this cohort but with a very heterologous distribution within the samples (Figure 4). Analogically to SLC2A3, despite of lack of significance in a group statistical analysis, WB showed a drastic reduction in the level of this protein in 11 (out of 13) seminal plasma samples from patient with NOA (Figure 4).
SMS is expressed in almost all human tissues but both at the mRNA and protein levels the most intense expression takes place in male and female reproductive organs (according to Human Protein Atlas). SMS catalyzes the production of spermine (from spermidine and decarboxylated S-adenosylmethionine) which is a polyamine involved in cellular metabolism in animals, plants, some fungi and bacteria (64). Experimental and clinical data clearly show that SMS is crucial for normal development in humans and other mammals; an experiment with transgenic mice showed that SMS is required for normal growth, viability and fertilization (65) whereas in humans, alterations in SMS gene have been connected with the X-linked recessive condition termed Snyder-Robinson syndrome (SRS) characterized with mild-to-moderate mental retardation, muscle and bone abnormalities, facial dysmorphism and other symptoms (66-68). Although SMS is widely expressed within eukaryotic cells, it is little known about this protein in context of male fertility. Male mice carrying a chromosomal deletion of part of the chromosome X containing most of the SMS gene had diminished numbers of meiotic and postmeiotic cells in their testes (65). Also, at the RNA level, seminal plasma SMS was found to be reduced in patients with asthenozoospermia (69). It can be concluded that local lack of spermine in testicular cells may be a cause of NOA in the patients under study although. According to the Human Protein Atlas, SMS is present in testes, epididymis and prostate but testes are the place where the protein is the most abundantly present.
There are several studies that applied proteomics to assess seminal plasma biomarkers in disrupted spermatogenesis. Some of them adapted separation and quantification of seminal plasma proteins in 2‑dimentional electrophoresis (2DE) before being selected for massspectrometry identification (8,70) while the others used more advanced mass-spectrometry based protein quantification (71,72). Our proteomic approach was based on relatively big group of samples (7–17 samples in a group, depending on the infertility type and analysis) and a robust spectra quantification method (25,26) selecting possible protein candidates indicative of male infertility. Looking at the protein-protein interaction analysis, it can be noticed that some of the proteins identified within one infertile group do not interact with other proteins suggesting the presence of different mechanisms (Figure 5). The selected in our study proteins are novel when compared to other proteomic studies aiming to identify infertility biomarkers (8,72,73). However, several of the identified proteins, namely LTF, SEMG1 and SEMG2 were already identified in seminal plasma from fertile donors in other studies and were referred as fertility-connected (23). The generated in our study list of differentially expressed proteins is rather short however, this outcome stands in a line with the outcomes of other studies aiming to identify specific biomarkers for male infertility (23,71).

Conclusions
In our study, we have tried to analyze and compare seminal plasma proteomes from infertile patients having problems with very affected sperm count suggesting severe problems with spermatogenesis. The application of high-throughput nanoUPLC-MS and the quantification approach eliminating the presence of false positives (25), have resulted in the identification of relatively small number of DEPs in males with SO, C and NOA (Table 1). Western immunoblotting performed for single individuals within a studied cohort provided a precise insight into the distribution of DEPs abolishing the limitations of MS analysis. The obtained data showed that the protein content among the samples studied was not unique (Figures 3,4). The most interesting candidate for a potential biomarker of spermatogenesis status, ADGRG2, turned out to be more attractive in context of C rather than azoospermia, whereas SLC2A3 and SMS are the proteins definitely requiring more attention regarding NOA studies. The presented data clearly suggest that different mechanisms can stand behind the same phenotypic entity of male infertility. It can be predicted that the knowledge on these mechanisms would lead in the future to the composition of specific protein panels useful for diagnostic and prognostic purposes in the treatment of male infertility while using non-invasive approach with seminal plasma protein content.
Acknowledgments
The nanoUPLC-MS/MS analysis was done in Mass Spectrometry Lab at the Institute of Biochemistry and Biophysics, Polish Academy of Sciences.
Funding: This work was supported by the National Science Centre (grant Nos. 2015/17/B/NZ2/01157 and 2020/37/B/NZ5/00549).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://tau.amegroups.com/article/view/10.21037/tau-23-130/rc
Data Sharing Statement: Available at https://tau.amegroups.com/article/view/10.21037/tau-23-130/dss
Peer Review File: Available at https://tau.amegroups.com/article/view/10.21037/tau-23-130/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tau.amegroups.com/article/view/10.21037/tau-23-130/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the National Bioethical Committee, Ministry of Health, Warsaw, Poland; code: OkB-5-2/15 (No.: OKB.078.6.2015) and informed consent was obtained from all individual participants.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Baker K, Sabanegh E Jr. Obstructive azoospermia: reconstructive techniques and results. Clinics (Sao Paulo) 2013;68:61-73. [Crossref] [PubMed]
- Tharakan T, Luo R, Jayasena CN, et al. Non-obstructive azoospermia: current and future perspectives. Fac Rev 2021;10:7. [Crossref] [PubMed]
- Camargo M, Intasqui P, Bertolla RP. Understanding the seminal plasma proteome and its role in male fertility. Basic Clin Androl 2018;28:6. [Crossref] [PubMed]
- De Lazari FL, Sontag ER, Schneider A, et al. Seminal plasma proteins and their relationship with sperm motility and morphology in boars. Andrologia 2019;51:e13222. [Crossref] [PubMed]
- Li Y, Sun Y, Ni A, et al. Seminal Plasma Proteome as an Indicator of Sperm Dysfunction and Low Sperm Motility in Chickens. Mol Cell Proteomics 2020;19:1035-46. [Crossref] [PubMed]
- Kasimanickam RK, Kasimanickam VR, Arangasamy A, et al. Sperm and seminal plasma proteomics of high- versus low-fertility Holstein bulls. Theriogenology 2019;126:41-8. [Crossref] [PubMed]
- Wang J, Wang J, Zhang HR, et al. Proteomic analysis of seminal plasma from asthenozoospermia patients reveals proteins that affect oxidative stress responses and semen quality. Asian J Androl 2009;11:484-91. [Crossref] [PubMed]
- Davalieva K, Kiprijanovska S, Noveski P, et al. Proteomic analysis of seminal plasma in men with different spermatogenic impairment. Andrologia 2012;44:256-64. [Crossref] [PubMed]
- Brown CO, Robbins BL, McKiernan HE, et al. Direct seminal fluid identification by protease-free high-resolution mass spectrometry. J Forensic Sci 2021;66:1017-23. [Crossref] [PubMed]
- Gaitskell-Phillips G, Martín-Cano FE, Ortiz-Rodríguez JM, et al. Seminal plasma proteins as potential biomarkers for sperm motility and velocities. Theriogenology 2022;177:34-41. [Crossref] [PubMed]
- Nowicka-Bauer K, Malcher A, Włoczkowska O, et al. Evaluation of seminal plasma HSPA2 protein as a biomarker of human spermatogenesis status. Reprod Biol 2022;22:100597. [Crossref] [PubMed]
- Yao L, Guo Y, Zhang X, et al. Quantitative proteomic biomarkers from extracellular vesicles of human seminal plasma in the differential diagnosis of azoospermia. Clin Transl Med 2021;11:e423. [Crossref] [PubMed]
- Azpiazu R, Amaral A, Castillo J, et al. High-throughput sperm differential proteomics suggests that epigenetic alterations contribute to failed assisted reproduction. Hum Reprod 2014;29:1225-37. [Crossref] [PubMed]
- S Al-Ouqaili MT. Detection of partial and/or complete Y chromosome microdeletions of azoospermia factor a (AZFa) sub-region in infertile Iraqi patients with azoospermia and severe oligozoospermia. J Clin Lab Anal 2022;36:e24272. [Crossref] [PubMed]
- Malcher A, Rozwadowska N, Stokowy T, et al. Potential biomarkers of nonobstructive azoospermia identified in microarray gene expression analysis. Fertil Steril 2013;100:1686-94.e1-7.
- Ali H, Unar A, Zubair M, et al. In silico analysis of a novel pathogenic variant c.7G > A in C14orf39 gene identified by WES in a Pakistani family with azoospermia. Mol Genet Genomics 2022;297:719-30. [Crossref] [PubMed]
- Wang X, Jin HR, Cui YQ, et al. Case study of a patient with cryptozoospermia associated with a recessive TEX15 nonsense mutation. Asian J Androl 2018;20:101-2. [Crossref] [PubMed]
- Miki K, Willis WD, Brown PR, et al. Targeted disruption of the Akap4 gene causes defects in sperm flagellum and motility. Dev Biol 2002;248:331-42. [Crossref] [PubMed]
- Fang X, Huang LL, Xu J, et al. Proteomics and single-cell RNA analysis of Akap4-knockout mice model confirm indispensable role of Akap4 in spermatogenesis. Dev Biol 2019;454:118-27. [Crossref] [PubMed]
- F Riesco M. ProAKAP4 as Novel Molecular Marker of Sperm Quality in Ram: An Integrative Study in Fresh, Cooled and Cryopreserved Sperm. Biomolecules 2020;10:1046. [Crossref] [PubMed]
- Légaré C, Cloutier F, Makosso-Kallyth S, et al. Cysteine-rich secretory protein 1 in seminal plasma: potential biomarker for the distinction between obstructive and nonobstructive azoospermia. Fertil Steril 2013;100:1253-60. [Crossref] [PubMed]
- Diamandis EP, Arnett WP, Foussias G, et al. Seminal plasma biochemical markers and their association with semen analysis findings. Urology 1999;53:596-603. [Crossref] [PubMed]
- Milardi D, Grande G, Vincenzoni F, et al. Proteomic approach in the identification of fertility pattern in seminal plasma of fertile men. Fertil Steril 2012;97:67-73.e1. [Crossref] [PubMed]
- Reiter L, Claassen M, Schrimpf SP, et al. Protein identification false discovery rates for very large proteomics data sets generated by tandem mass spectrometry. Mol Cell Proteomics 2009;8:2405-17. [Crossref] [PubMed]
- Bakun M, Karczmarski J, Poznanski J, et al. An integrated LC-ESI-MS platform for quantitation of serum peptide ladders. Application for colon carcinoma study. Proteomics Clin Appl 2009;3:932-46. [Crossref] [PubMed]
- Malinowska A, Kistowski M, Bakun M, et al. Diffprot - software for non-parametric statistical analysis of differential proteomics data. J Proteomics 2012;75:4062-73. [Crossref] [PubMed]
- World Health Organization. WHO laboratory manual for the examination and processing of human semen. Fifth ed. 2010;287.
- Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods 2007;4:207-14. [Crossref] [PubMed]
- Dietrich MA, Judycka S, Żarski D, et al. Proteomic analysis of pikeperch seminal plasma provides novel insight into the testicular development of domesticated fish stocks. Animal 2021;15:100279. [Crossref] [PubMed]
- Banaś AM, Bocian-Ostrzycka KM, Dunin-Horkawicz S, et al. Interplay between DsbA1, DsbA2 and C8J_1298 Periplasmic Oxidoreductases of Campylobacter jejuni and Their Impact on Bacterial Physiology and Pathogenesis. Int J Mol Sci 2021;22:13451. [Crossref] [PubMed]
- Nowicka-Bauer K, Lepczynski A, Ozgo M, et al. Sperm mitochondrial dysfunction and oxidative stress as possible reasons for isolated asthenozoospermia. J Physiol Pharmacol 2018; [PubMed]
- Obermann H, Samalecos A, Osterhoff C, et al. HE6, a two-subunit heptahelical receptor associated with apical membranes of efferent and epididymal duct epithelia. Mol Reprod Dev 2003;64:13-26. [Crossref] [PubMed]
- Suchý T, Zieschang C, Popkova Y, et al. The repertoire of Adhesion G protein-coupled receptors in adipocytes and their functional relevance. Int J Obes (Lond) 2020;44:2124-36. [Crossref] [PubMed]
- Balenga N, Azimzadeh P, Hogue JA, et al. Orphan Adhesion GPCR GPR64/ADGRG2 Is Overexpressed in Parathyroid Tumors and Attenuates Calcium-Sensing Receptor-Mediated Signaling. J Bone Miner Res 2017;32:654-66. [Crossref] [PubMed]
- Grunddal KV, Tonack S, Egerod KL, et al. Adhesion receptor ADGRG2/GPR64 is in the GI-tract selectively expressed in mature intestinal tuft cells. Mol Metab 2021;51:101231. [Crossref] [PubMed]
- Davies B, Behnen M, Cappallo-Obermann H, et al. Novel epididymis-specific mRNAs downregulated by HE6/Gpr64 receptor gene disruption. Mol Reprod Dev 2007;74:539-53. [Crossref] [PubMed]
- Watanabe H, Takeo T, Tojo H, et al. Lipocalin 2 binds to membrane phosphatidylethanolamine to induce lipid raft movement in a PKA-dependent manner and modulates sperm maturation. Development 2014;141:2157-64. [Crossref] [PubMed]
- Yin Q, Shen J, Wan X, et al. Impaired sperm maturation in conditional Lcn6 knockout mice. Biol Reprod 2018;98:28-41. [Crossref] [PubMed]
- Zhao Y, Diao H, Ni Z, et al. The epididymis-specific antimicrobial peptide β-defensin 15 is required for sperm motility and male fertility in the rat (Rattus norvegicus). Cell Mol Life Sci 2011;68:697-708. [Crossref] [PubMed]
- Zhang DL, Sun YJ, Ma ML, et al. Gq activity- and β-arrestin-1 scaffolding-mediated ADGRG2/CFTR coupling are required for male fertility. Elife 2018;7:e33432. [Crossref] [PubMed]
- Davies B, Baumann C, Kirchhoff C, et al. Targeted deletion of the epididymal receptor HE6 results in fluid dysregulation and male infertility. Mol Cell Biol 2004;24:8642-8. [Crossref] [PubMed]
- Tan MQ, Huang WJ, Lan FH, et al. Genetic mutation analysis of 22 patients with congenital absence of vas deferens: a single-center study. Biol Reprod 2022;106:108-17. [Crossref] [PubMed]
- Pagin A, Bergougnoux A, Girodon E, et al. Novel ADGRG2 truncating variants in patients with X-linked congenital absence of vas deferens. Andrology 2020;8:618-24. [Crossref] [PubMed]
- Yuan P, Liang ZK, Liang H, et al. Expanding the phenotypic and genetic spectrum of Chinese patients with congenital absence of vas deferens bearing CFTR and ADGRG2 alleles. Andrology 2019;7:329-40. [Crossref] [PubMed]
- Wang H, An M, Liu Y, et al. Genetic diagnosis and sperm retrieval outcomes for Chinese patients with congenital bilateral absence of vas deferens. Andrology 2020;8:1064-9. [Crossref] [PubMed]
- Khan MJ, Pollock N, Jiang H, et al. X-linked ADGRG2 mutation and obstructive azoospermia in a large Pakistani family. Sci Rep 2018;8:16280. [Crossref] [PubMed]
- Bieth E, Hamdi SM, Mieusset R. Genetics of the congenital absence of the vas deferens. Hum Genet 2021;140:59-76. [Crossref] [PubMed]
- Raffaniello RD. Rab3 proteins and cancer: Exit strategies. J Cell Biochem 2021;122:1295-301. [Crossref] [PubMed]
- Shan MM, Sun SC. The multiple roles of RAB GTPases in female and male meiosis. Hum Reprod Update 2021;27:1013-29. [Crossref] [PubMed]
- Manabe S, Nishimura N, Yamamoto Y, et al. Identification and characterization of Noc2 as a potential Rab3B effector protein in epithelial cells. Biochem Biophys Res Commun 2004;316:218-25. [Crossref] [PubMed]
- Sidhu RS, Bhullar RP. Rab3B in human platelet is membrane bound and interacts with Ca(2+)/calmodulin. Biochem Biophys Res Commun 2001;289:1039-43. [Crossref] [PubMed]
- Pearl CA, Roser JF. Expression of lactoferrin in the boar epididymis: effects of reduced estrogen. Domest Anim Endocrinol 2008;34:153-9. [Crossref] [PubMed]
- Wichmann L, Vaalasti A, Vaalasti T, et al. Localization of lactoferrin in the male reproductive tract. Int J Androl 1989;12:179-86. [Crossref] [PubMed]
- Wang P, Liu B, Wang Z, et al. Characterization of lactoferrin receptor on human spermatozoa. Reprod Biomed Online 2011;22:155-61. [Crossref] [PubMed]
- Hamada A, Sharma R, du Plessis SS, et al. Two-dimensional differential in-gel electrophoresis-based proteomics of male gametes in relation to oxidative stress. Fertil Steril 2013;99:1216-1226.e2. [Crossref] [PubMed]
- Sharma R, Agarwal A, Mohanty G, et al. Proteomic analysis of seminal fluid from men exhibiting oxidative stress. Reprod Biol Endocrinol 2013;11:85. [Crossref] [PubMed]
- Soudmand P, Tofighi A, Tolouei Azar J, et al. Different continuous exercise training intensities induced effect on sertoli-germ cells metabolic interaction; implication on GLUT-1, GLUT-3 and MCT-4 transporting proteins expression level. Gene 2021;783:145553. [Crossref] [PubMed]
- Zhang LL, Ma J, Yang B, et al. Interference with lactate metabolism by mmu-miR-320-3p via negatively regulating GLUT3 signaling in mouse Sertoli cells. Cell Death Dis 2018;9:964. [Crossref] [PubMed]
- Noveski P, Popovska-Jankovic K, Kubelka-Sabit K, et al. MicroRNA expression profiles in testicular biopsies of patients with impaired spermatogenesis. Andrology 2016;4:1020-7. [Crossref] [PubMed]
- Oliveira PF, Alves MG, Rato L, et al. Influence of 5α-dihydrotestosterone and 17β-estradiol on human Sertoli cells metabolism. Int J Androl 2011;34:e612-20. [Crossref] [PubMed]
- Rato L, Alves MG, Socorro S, et al. Metabolic modulation induced by oestradiol and DHT in immature rat Sertoli cells cultured in vitro. Biosci Rep 2012;32:61-9. [Crossref] [PubMed]
- Sarkar D, Singh SK. Neonatal hypothyroidism affects testicular glucose homeostasis through increased oxidative stress in prepubertal mice: effects on GLUT3, GLUT8 and Cx43. Andrology 2017;5:749-62. [Crossref] [PubMed]
- Liu X, Liu F. In depth mapping of human testicular and epididymal proteins and their functional association with spermatozoa. Mol Med Rep 2015;12:173-9. [Crossref] [PubMed]
- Pegg AE, Michael AJ. Spermine synthase. Cell Mol Life Sci 2010;67:113-21. [Crossref] [PubMed]
- Wang X, Ikeguchi Y, McCloskey DE, et al. Spermine synthesis is required for normal viability, growth, and fertility in the mouse. J Biol Chem 2004;279:51370-5. [Crossref] [PubMed]
- Marhabaie M, Hickey SE, Miller K, et al. Maternal mosaicism for a missense variant in the SMS gene that causes Snyder-Robinson syndrome. Cold Spring Harb Mol Case Stud 2021;7:a006122. [Crossref] [PubMed]
- Qazi TJ, Wu Q, Aierken A, et al. Whole-exome sequencing identifies a novel mutation in spermine synthase gene (SMS) associated with Snyder-Robinson Syndrome. BMC Med Genet 2020;21:168. [Crossref] [PubMed]
- Larcher L, Norris JW, Lejeune E, et al. The complete loss of function of the SMS gene results in a severe form of Snyder-Robinson syndrome. Eur J Med Genet 2020;63:103777. [Crossref] [PubMed]
- Chen L, Wen CW, Deng MJ, et al. Metabolic and transcriptional changes in seminal plasma of asthenozoospermia patients. Biomed Chromatogr 2020;34:e4769. [Crossref] [PubMed]
- Yamakawa K, Yoshida K, Nishikawa H, et al. Comparative analysis of interindividual variations in the seminal plasma proteome of fertile men with identification of potential markers for azoospermia in infertile patients. J Androl 2007;28:858-65. [Crossref] [PubMed]
- Rolland AD, Lavigne R, Dauly C, et al. Identification of genital tract markers in the human seminal plasma using an integrative genomics approach. Hum Reprod 2013;28:199-209. [Crossref] [PubMed]
- Drabovich AP, Jarvi K, Diamandis EP. Verification of male infertility biomarkers in seminal plasma by multiplex selected reaction monitoring assay. Mol Cell Proteomics 2011;10:M110.004127.
- Cui Z, Agarwal A, da Silva BF, et al. Evaluation of seminal plasma proteomics and relevance of FSH in identification of nonobstructive azoospermia: A preliminary study. Andrologia 2018;50:e12999. [Crossref] [PubMed]