
Silencing SPP1 in M2 macrophages inhibits the progression of castration-resistant prostate cancer via the MMP9/TGFβ1 axis
Highlight box
Key findings
• This study proposes that the high expression of secreted phosphoprotein 1 (SPP1) in M2 macrophages can promote epithelial-mesenchymal transition in castration-resistant prostate cancer (CRPC) cells through the matrix metallopeptidase 9 (MMP9)/transforming growth factor beta 1 (TGFβ1) signaling pathway, thereby enhancing the progression of CRPC.
What is known and what is new?
• M2 macrophages play an important role in the progression of CRPC.
• It is still unclear whether the high expression of SPP1 in M2 macrophages is an important factor promoting the progression of CRPC.
What is the implication, and what should change now?
• This research result suggests that targeted therapy for SPP1 has the potential to inhibit the tumor promoting effect of M2 macrophages, thereby inhibiting the progression of prostate cancer.
Introduction
Prostate cancer (PCa) is a common malignant tumor of the urinary system in men, and its incidence and mortality rates have been steadily increasing in Western countries (1). Following prolonged androgen deprivation therapy (ADT), the majority of PCa patients inevitably progress to castration-resistant prostate cancer (CRPC) (2). Recent research has found that in CRPC patients with bone metastasis, M2 macrophages are the main cellular population in the tumor microenvironment (TME), indicating that tumor-associated macrophages (TAMs) may play a crucial role in mediating the progression of CRPC (3). However, the specific mechanisms by which macrophages influence the progression of PCa remain unclear.
TME forms the foundation for tumor metastasis with its complex cellular components. Cell populations commonly found within this microenvironment, such as fibroblasts and macrophages, play a crucial role in tumor progression (4,5). Targeted therapies focusing on relevant cells within the TME have shown promise in inhibiting tumor progression (6,7). PCa is known for its cold immune response to immunotherapy. The main reason is that the relevant immune cells are unable to heavily infiltrate the TME of PCa (8). Nevertheless, recent research has revealed a high abundance of macrophages in CRPC tissues (3). In addition, a high abundance of macrophages can suppress T cell activity, leading to an immunosuppressive microenvironment that promotes castration-resistant progression of PCa (9). Thus, targeting macrophages has emerged as a potential approach for PCa treatment (10).
One important molecule of interest is secreted phosphoprotein 1 (SPP1), also known as osteopontin, which is a multifunctional secreted phosphorylated glycoprotein (11). SPP1 is primarily expressed in bone-forming cells, fibroblasts, macrophages, dendritic cells, lymphocytes, and monocytes within the immune system (12). Research has demonstrated a high expression of SPP1 in solid organ tumors (13). Furthermore, SPP1 is also considered a biomarker of M2 macrophages, and recent research indicates that the ratio of CXCL9/SPP1 expression in macrophages can serve as a marker for measuring macrophage polarization (11,14). SPP1 in macrophages has been shown to promote tumor metastasis and malignant progression in various types of cancer (14-17). Ramadan et al. found that continuous oral administration of 100 mg/kg thioacetamide (TAA) significantly inhibited the expression of SPP1 in liver fibrosis tissues (18). However, the potential of SPP1 to promote tumor metastasis within macrophages has not yet been explored in PCa research.
In our study, analysis of the GSE32269 dataset revealed that SPP1 exhibited the highest differential expression in metastatic CRPC (mCRPC) tissues compared to localized PCa. Additionally, we found that SPP1 is mainly expressed in mononuclear macrophages in the PCa single-cell RNA sequencing (scRNA-seq) database. Through in vitro and in vivo experiments, it was confirmed that high expression of SPP1 in M2 macrophages promotes PCa metastasis. Mechanistic studies have elucidated a robust correlation between SPP1 and matrix metallopeptidase 9 (MMP9), highlighting that SPP1 regulates the expression of MMP9 in M2 macrophages through the PI3K/AKT signaling pathway. MMP9 is known to activate the extracellular secretion of transforming growth factor beta 1 (TGFβ1) by cleaving TGFβ1 protein (19). In summary, our study provides evidence that high expression of SPP1 in M2 macrophages is an important factor in promoting PCa metastasis, and SPP1 exerts its effects through the regulation of MMP9/TGFβ1. We present this article in accordance with the MDAR and ARRIVE reporting checklists (available at https://tau.amegroups.com/article/view/10.21037/tau-24-127/rc).
Methods
Bioinformatics analysis
In the Gene Expression Omnibus (GEO) database, we analyzed the transcriptomic sequencing data of GSE32269 and performed a volcano plot analysis. In the PCa database (https://bioinformatics.cruk.cam.ac.uk/apps/camcAPP/), we analyzed the relationship between the expression of SPP1 and the progression of PCa. The UALCAN (https://ualcan.path.uab.edu/index.html) and GEPIA (http://gepia.cancer-pku.cn/index.html) databases were used to analyze the association of SPP1 expression with Gleason score, tumor-node-metastasis (TNM) stage, and patient survival in PCa. The single-cell database TISCH2 (http://tisch.comp-genomics.org/) was utilized to analyze the cell clustering of different types of PCa and the cellular distribution of SPP1. The Timer2 (http://timer.cistrome.org/) and cBioPortal (https://www.cbioportal.org/) databases were employed to analyze the expression of SPP1 in PCa and its relationship with M1/M2 macrophages. The Kyoto Encyclopedia of Genes and Genomes (KEGG) and gene set enrichment analysis (GSEA) enrichment were performed by the OECloud tools at https://cloud.oebiotech.com. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Cell culture
The human PCa cell lines PC3 and the human monocytic leukemia cell line THP-1, as well as the mouse PCa cell line RM1 were obtained from American Type Culture Collection (ATCC; Manassas, VA, USA). All four cell lines were cultured in high-glucose Dulbecco’s modified Eagle medium (DMEM; Gibco, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (FBS; Gibco) and 1% penicillin/streptomycin. However, THP-1 did not require the addition of 1% penicillin/streptomycin. The cells were maintained at 37 °C in a humidified atmosphere containing 0.5% CO2. Before experimentation, all used cell lines were authenticated through short tandem repeat (STR) analysis and were confirmed to be free of mycoplasma contamination.
Cell transfection
Three specific small interfering RNA (siRNA) targeting SPP1 were designed and constructed by Beijing Tsingke Biotech Co., Ltd. THP-1 cells were seeded in a 6-well plate and induced with 100 ng/mL of phorbol myristate acetate (PMA) [MedChemExpress (MCE), Cat#HY-18739; MCE, Monmouth Junction, NJ, USA] to generate M0 macrophages, followed by induction with IL-4 (MCE, Cat#HY-P70445) to generate M2 macrophages. Subsequently, Lipo3000 transfection reagent (Thermo Fisher Scientific, Waltham, MA, USA) was used for siRNA transfection of the cells. The siRNA sequences used are shown in Table 1.
Table 1
| Gene | Sense | Anti-sense |
|---|---|---|
| SPP1-1 | CCAAAGUCAGCCGUGAAUU | AAUUCACGGCUGACUUUGG |
| SPP1-2 | GUAAGGAAGAAGAUAAACA | UGUUUAUCUUCUUCCUUAC |
| SPP1-3 | GGUCAAAAUCUAAGAAGUU | AACUUCUUAGAUUUUGACC |
| MMP9 | CAAGACAAAGCCUAUUUCUTT | AGAAAUAGGCUUUGUCUUGTT |
siRNA, small interfering RNA.
Western blot
Cellular proteins were extracted using radioimmunoprecipitation assay (RIPA) lysis buffer and quantified using a bicinchoninic acid (BCA) assay kit according to the manufacturer’s protocol. Then, 2 micrograms of protein were loaded onto sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) gels and separated by electrophoresis. The proteins were then transferred to immune-blot polyvinylidene fluoride (PVDF) membranes using a semi-dry transfer apparatus (Bio-Rad, Hercules, CA, USA). The membranes were blocked with a protein-free rapid sealing solution (#G2052, Servicebio, Wuhan, China) for 10 minutes at room temperature, followed by overnight incubation at 4 °C with primary antibodies. After washing the membranes three times for 15 minutes each with phosphate-buffered saline with Tween 20 (PBST), they were incubated at room temperature for 1 hour with secondary antibodies specific to the species. The membranes were then washed 3 times with PBST for 15 minutes each time. Protein signal detection was performed using the Ultrasensitive ECL Western HRP Substrate (#17047, ZenBio, Shanghai, China) and a Bio-Rad ChemiDoc MP System (170–8280). The relevant information on the antibodies used in the experiment is shown in Table 2.
Table 2
| Antibodies | Company | Catalog No. | Species | Molecular weight (kDa) |
|---|---|---|---|---|
| SPP1 | Proteintech | 22952-1-AP | Rabbit | 105 |
| MMP9 | Proteintech | 10375-2-AP | Rabbit | 78 |
| AGR1 | Proteintech | 66129-1-Ig | Mouse | 36 |
| TNFα | Proteintech | 26405-1-AP | Rabbit | 26 |
| CDH1 | Proteintech | 60335-1-Ig | Mouse | 120 |
| CDH2 | Proteintech | 66219-1-Ig | Mouse | 130 |
| Vimentin | Proteintech | 60330-1-Ig | Mouse | 57 |
| SNAIL | Proteintech | 13099-1-AP | Rabbit | 33 |
| PI3K | Zenbio | R381065 | Rabbit | 56 |
| p-PI3K | Zenbio | 341468 | Rabbit | 58 |
| AKT | Zenbio | R23412 | Rabbit | 54 |
| p-AKT | Zenbio | R381555 | Rabbit | 54 |
| Smad2/3 | Zenbio | 382472 | Rabbit | 52 |
| p-Smad2/3 | Zenbio | 251795 | Rabbit | 52 |
| β-actin | Zenbio | T200068-8F10 | Mouse | 42 |
SPP1, secreted phosphoprotein 1; MMP9, matrix metallopeptidase 9; TNF, tumor necrosis factor.
Real-time quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted using a TRIzol reagent (Ambion 317908), and the concentration was determined. The RNA was then reverse transcribed into complementary DNA (cDNA) using PrimeScript Reverse Transcriptase (DRR047A, TaKaRa, Beijing, China). For gene expression analysis, SYBR Green Master Mix (A25742, China) from Thermo Fisher Scientific and primers synthesized by Sangon Biotech (Shanghai, China) were used. RT-qPCR was performed on a Bio-Rad CFX Connect Real-Time System. The gene expression levels were determined using the 2−ΔΔCt method.
Construction of luciferase labeled cells
The RM1 mouse PCa cell line was seeded into a 24-well plate until reaching approximately 75% confluency. Then, 10 µL of blank plasmid lentiviral particles provided by Beijing Tsingke Biotech Co., Ltd. were added and mixed well. After 24 hours, the media was replaced and supplemented with puromycin for selection until stable cells carrying the luciferase fluorescent label were obtained.
Orthotopic PCa model and bioluminescence imaging (BLI)
All animal studies carried out in this project strictly followed the regulations of the China Nature Conservation Committee and were approved by the Animal Experiment Welfare and Ethics Committee of the Army Medical University (No. AMUWEC20232943), in compliance with institutional guidelines for the care and use of animals. A protocol was prepared before the study without registration. A total of eight C57 male mice aged 4–6 weeks were purchased from the Animal Research Institute of the Third Military Medical University, and four were randomly selected and divided into an experimental group and a control group, respectively. The mice were raised in a sterile specific-pathogen-free (SPF) environment, and the feed and water provided were also treated aseptically. RM1-luciferase cells (1×105) were resuspended in 10–20 µL phosphate-buffered saline (PBS) and injected into the prostate of the mice using an insulin needle under a microscope after opening the abdominal cavity and exposing the prostate gland. The surgical site was closed with 4-0 silk thread, and antibiotics were added to the drinking water to prevent infection. BLI, stimulated by D-luciferin potassium salt solution (10 µL/g body weight, #ST198, Beyotime, China), was performed post-surgery using the IVIS Spectrum CT system (PerkinElmer, Waltham, MA, USA) provided by the First Affiliated Hospital of the Third Military Medical University. Subsequently, 0.5% sodium carboxymethyl cellulose (CMC Na) and 0.5% CMC-TAA was administered orally at a dose of 100 mg/kg/day for 15 consecutive days. On the last day, the mice were euthanized and prostate tumor tissue was extracted.
Tibial bone metastasis model
A total of eight C57 mice aged 4–6 weeks were randomly divided into an experimental group and a control group at 5–6 weeks of age, with four mice in each group. The mice were anesthetized with isoflurane gas and the left hind limb was disinfected with alcohol. Using an insulin syringe, 1×105 of RM1-luciferase cells were drawn up and resuspended in 20 µL PBS. The insulin needle was slowly inserted vertically along the tibial plateau into the bone marrow cavity for injection, followed by disinfection of the injection site. On the third day after injection, live imaging was performed to detect the fluorescence of the tibia and ensure consistent cell injection. Subsequently, 0.5% CMC-Na and 0.5% CMC-TAA was administered orally at a dose of 100 mg/kg/day for 15 consecutive days. On day 33, the mice were euthanized and their tibias collected, which were fixed in 4% paraformaldehyde for at least 24 hours after decalcification. Relevant indicators were evaluated by hematoxylin and eosin (H&E) staining, immunohistochemical (IHC) staining, and tartrate-resistant acid phosphatase (TRAP) staining.
Immunohistochemistry and immunofluorescence
The paraffin-embedded PCa tumor tissue was sectioned into 5-mm thick slices. Ethylenediamine tetraacetic acid (EDTA) retrieval was performed at a high temperature for 3 minutes, followed by PBS washing and serum blocking for 1 hour. The corresponding primary antibody was added under dark conditions and incubated overnight at 4 °C. After PBS washing the next day, the secondary antibody was added and incubated at room temperature for 30 minutes. Immunohistochemistry was performed using 3,3'-diaminobenzidine (DAB) chromogen to visualize the staining. Subsequently, the slides were placed in a hematoxylin solution for 1 minute, rinsed with running water, and air-dried before being covered and slipped. The immunofluorescence steps were conducted the same as for immunohistochemistry, with the addition of 4',6-diamidino-2-phenylindole (DAPI) for nuclear staining. Finally, the slides were sealed with a mounting medium containing an anti-fluorescent quencher.
Flow cytometry
After washing the treated cells with PBS, they were resuspended in 200 µL of PBS and incubated with flow cytometry antibodies according to the appropriate proportions. The antibodies included anti-human CD68 antibody (#11-0689-41, Thermo Fisher Scientific, ASA), anti-human CD163 (#17-1639-41, Thermo Fisher Scientific), and anti-human CD86 (#12-0869-41, Thermo Fisher Scientific) antibodies. M0 macrophages were identified as CD68+, M1 macrophages were identified as CD68+CD86+, and M2 macrophages were identified as CD68+CD163+. The experimental detection instrument (BD LSRFortessaTM; Becton, Dickinson, and Co., Franklin Lakes, NJ, USA) was provided by the First Affiliated Hospital of the Third Military Medical University.
Statistical analysis
All data presented in this study are representative of at least three independent replicates. Quantitative data were expressed as the mean ± standard deviation and were analyzed using GraphPad Prism 8 software (GraphPad Software, San Diego, CA, USA). Differences between the two groups were compared using unpaired t-tests. A P value <0.05 was considered statistically significant.
Results
Upregulation of SPP1 is positively correlated with the progression of PCa
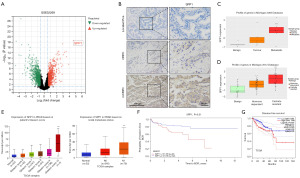
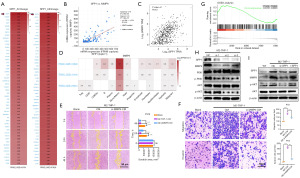
To investigate the potential mechanisms underlying CRPC metastasis, we analyzed the GSE32269 dataset containing transcriptomic sequencing data from tumor tissues of 21 patients with localized PCa and mCRPC. Our analysis revealed that SPP1 had the highest differential expression in the mCRPC group (Figure 1A). IHC analysis of tumor samples collected from patients at different stages of PCa progression indicated that SPP1 expression was lower in localized PCa but increased in CRPC tissues. Moreover, the highest expression of SPP1 was observed in tumor samples from mCRPC patients (Figure 1B). Additionally, the Michigan 2005 and 2012 databases showed higher expression levels of SPP1 in tumor samples from patients with mCRPC than in benign and localized PCa (Figure 1C). Analysis of The Cancer Genome Atlas (TCGA) database further revealed a positive correlation between SPP1 expression and the Gleason score, as well as TNM analysis in clinical PCa patients (Figure 1D,1E). Furthermore, we observed that patients with high expression of SPP1 had significantly shorter overall survival in the Memorial Sloan Kettering Cancer Center (MSKCC) and TCGA-PRAD (prostate adenocarcinoma) sequencing data (Figure 1F,1G). These findings suggest that the high expression of SPP1 in PCa tissue is associated with the malignant progression of PCa.
SPP1 is predominantly expressed in macrophages in PCa tissues
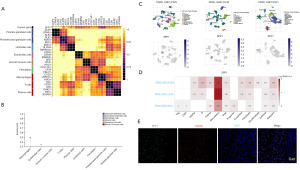
Based on our analysis of the GSE32269 dataset through transcriptomic sequencing, we observed a significantly high expression of SPP1 in mCRPC tumor tissues, indicating a substantial differential fold change. However, due to the intricate cellular composition within the TME, it remained challenging to identify the predominant cell type responsible for SPP1 expression. To address this, we initially examined the Human Protein Atlas (HPA) database to analyze the cellular components associated with SPP1. Interestingly, our findings revealed a strong correlation between SPP1 and macrophage markers (C1QA, LRRC25, VSIG4) in the prostate (Figure 2A,2B). Moreover, when investigating three PCa single-cell datasets (GSE137829, GSE141445, GSE176031) in the TISHC2 tumor single-cell database, we discovered that SPP1 is primarily expressed in monocyte-derived macrophages, whereas its expression in other cell types is minimal (Figure 2C,2D). Additionally, immunofluorescence staining performed on tumor tissues from clinical PCa patients exhibited co-localization of SPP1 and CD206 in mCRPC tumor tissues (Figure 2E). Taken together, these findings suggest that SPP1 is primarily expressed in M2 macrophages within PCa.
The plasticity regulation of macrophage polarization is mediated by the expression of SPP1
Macrophages are a prominent component of the complex PCa microenvironment. To explore the relationship between SPP1 and macrophage polarization, we analyzed the correlation between SPP1 and M1/M2 macrophage polarization markers in the SU2C-mCRPC dataset. Our findings revealed a significantly higher correlation between SPP1 and M2 macrophage markers (CD206, CD115, CD163, IL-10, CXCL3, CCL18) compared to M1 markers (TNFα, NOS2, CXCL10, CD86, CD80) (Figure 3A,3B). Moreover, analysis of the PCa immune infiltration database indicated that high SPP1 expression is more strongly associated with M2 macrophage infiltration than with M1 and M0 macrophages (Figure 3C). Furthermore, SPP1 was significantly upregulated in M2-polarized macrophages (Figure 3D). Knockdown of SPP1 in M2 macrophages resulted in decreased ARG1 expression and increased TNFα expression (Figure 3E). Flow cytometry analysis of M1/M2 markers demonstrated that knockdown of SPP1 in M2 macrophages led to a decrease in the proportion of CD68+CD163+ cells and an increase in the proportion of CD68+CD86+ cells (Figure 3F). Collectively, these results indicate that the expression level of SPP1 plays a crucial role in regulating macrophage polarization plasticity.
SPP1 in M2 macrophages promotes the progression of PCa
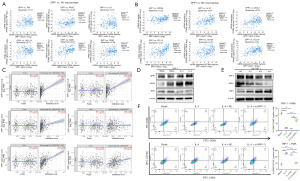
To confirm the role of SPP1 in M2 macrophages in promoting PCa metastasis, we collected conditioned media from M0, M1, and M2 macrophages with knocked-down SPP1 and co-cultured them with PC3 cells. Our results showed that the conditioned media from M2 macrophages significantly promoted the migration of PC3 cells compared to those from M0 macrophages. However, the migration capability of PC3 cells was significantly weakened when cultured in conditioned media from M2 macrophages with knocked-down SPP1 (Figure 4A). Similarly, Transwell assays demonstrated a substantial increase in the invasive and migratory capabilities of PC3 cells when exposed to conditioned media from M2 macrophages. However, this effect was attenuated when SPP1 was knocked down (Figure 4B). These findings demonstrate that the expression of SPP1 in M2 macrophages significantly influences the invasion and migration of PC3 cells.
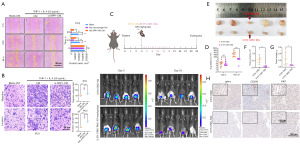
To validate the function of SPP1 in vivo, we implanted luciferase-labeled RM1 mouse PCa cells into the orthotopic prostate of C57BL mice. The experimental group was treated with the SPP1 inhibitor TAA at a dose of 100 mg/kg/day. At 3 days after implantation, similar fluorescence values were observed between the control group and the inhibitor group in the orthotopic prostate. However, on day 33, it was found that the fluorescence intensity in the inhibitor group was significantly reduced (Figure 4C,4D). By measuring the volume and weight of isolated prostate tumors, we found that the inhibitor-treated group exhibited a significant reduction in the size and weight of PCa tumors compared to the control group (Figure 4E-4G). IHC staining results showed a significant downregulation of SPP1 expression and a marked decrease in the number of M2 macrophages in the inhibitor-treated group, resulting in significant inhibition of tumor progression (Figure 4H). These findings provide further evidence that SPP1 in M2 macrophages mediates the progression of PCa.
Inhibiting the expression of SPP1 in macrophages significantly reduces tumor metastasis and bone destruction
In the mouse model of orthotopic PCa, the SPP1 inhibitor group exhibited significant suppression of tumor proliferation. To assess orthotopic metastasis of PCa, in vivo imaging tests were conducted 33 days after orthotopic implantation of PCa cells in mice. In the control group, a portion of mice showed orthotopic metastasis of PCa, specifically liver and spleen metastasis (Figure 5A). In contrast, no metastatic lesions were observed in any locations other than the prostate site in the inhibitor group (Figure 5B). Furthermore, the expression of the epithelial marker CDH1 was upregulated, while the mesenchymal markers CDH2, SNAIL, and vimentin were significantly downregulated in the CMC-TAA treat group (Figure 5C).
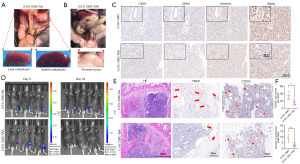
To evaluate tibial tumor metastasis, a mouse tibial tumor model was established, and tibial fluorescence was detected on days 3 and 33. The live imaging results demonstrated that tumor proliferation in the inhibitor-treated mice was significantly inhibited, with some tumors no longer exhibiting fluorescence (Figure 5D). Subsequent histopathological examination, including H&E staining, TRAP staining, and IHC staining, revealed a noteworthy reduction in bone trabecular destruction in the inhibitor group. Moreover, the number of osteoclasts and CD206+ macrophages was remarkably decreased compared to the control group (Figure 5E). The TRAP cells (indicated by red arrows) and CD206+ cells (indicated by red arrows) were statistically analyzed and presented (Figure 5F). These results collectively demonstrate that inhibiting SPP1 expression in M2 macrophages can significantly impede PCa metastasis.
SPP1 regulates the expression of MMP9 in M2 macrophages through the activation of the PI3K/AKT signaling pathway
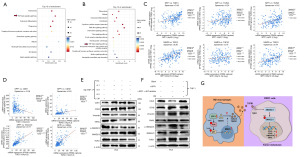
To investigate the specific mechanisms underlying the role of SPP1 in PCa progression, we analyzed SPP1-associated genes in two PCa single-cell datasets, namely GSE143791 and GSE137829. Among these genes, MMP9 exhibited the highest correlation with SPP1 (Figure 6A). Further analysis of the SU2C-mCRPC dataset revealed that MMP9 ranked second in terms of its correlation with SPP1, with a Spearman correlation coefficient of 0.79 (Figure 6B). Moreover, a significant correlation between SPP1 and MMP9 was confirmed in the TCGA-PRAD database (Figure 6C). Interestingly, the PCa single-cell dataset indicated that MMP9 is primarily expressed in macrophages (Figure 6D).
To validate the impact of MMP9 on PCa progression, we conducted scratch assays and Transwell experiments using conditioned media from MMP9-knockdown M2 macrophages. The results confirmed that the invasion and migration of PC3 cells were significantly inhibited (Figure 6E,6F). This suggests that SPP1 in M2 macrophages regulates the expression of MMP9, thereby promoting PCa progression.
To further explore the relationship between SPP1 and MMP9, we analyzed the genes associated with MMP9 in the GSE32269 dataset and performed GSEA. The results revealed a close association between MMP9 and the PI3K/AKT signaling pathway (Figure 6G). Given that SPP1 is a phosphorylated protein and both SPP1 and MMP9 are expressed in macrophages in PCa, we found that knocking down SPP1 in M2 macrophages led to reduced MMP9 expression and inhibition of the PI3K/AKT signaling pathway (Figure 6H). Additionally, by adding recombinant SPP1 to SPP1-knockdown cells, we observed upregulated MMP9 expression and restored phosphorylated AKT protein expression (Figure 6I). These findings suggest that SPP1 regulates MMP9 expression through the activation of the PI3K/AKT signaling pathway.
MMP9 facilitates the EMT of PC3 cells by promoting the processing of TGFβ through the activation of the SMAD2/3 pathway
Previous research has shown that the matrix metalloproteinase (MMP) family can cleave the latency-associated peptide (LAP) of TGF-β, leading to its activation and subsequent release of TGF-β ligands (20). Intriguingly, KEGG pathway enrichment analysis of differentially expressed genes co-expressed with SPP1 and MMP9 in the SU2C-mCRPC database revealed an enrichment of the TGFβ signaling pathway in both cases (Figure 7A,7B). In the mCRPC database, correlation analysis between the three molecules of the TGFβ family (TGFβ1, TGFβ2, TGFβ3) revealed that both SPP1 and MMP9 had the highest correlation with TGFβ1 (Figure 7C). Further analysis of the mCRPC database confirmed the ability of TGFβ1 to promote EMT by revealing the correlation between TGFβ1 and EMT-related molecules (CDH1, CDH2, vimentin, SNAIL) (Figure 7D). We verified that conditioned medium from SPP1-knockdown M2 macrophages led to downregulation of phosphorylated SMAD2/3 and stromal markers CDH2, vimentin, and SNAIL, whereas the epithelial marker CDH1 was upregulated in PC3 cells (Figure 7E). Subsequently, we added recombinant SPP1 and recombinant SPP1 protein along with an AKT phosphorylation inhibitor to THP-1 cells and co-cultured them with PC3 cells. The results showed that the SMAD2/3 signaling pathway was activated in the group treated with recombinant SPP1 protein, whereas it was deactivated in the group treated with the AKT inhibitor (Figure 7F). The schematic diagram of this study is as follows: Overexpression of SPP1 in M2 macrophages activates the PI3K/AKT signaling pathway, promoting the expression of MMP9. Subsequently, MMP9 cleaves TGFβ1, leading to its maturation and increased secretion, which binds to receptors on PC3 cells, activating downstream SMAD2/3 signaling. This ultimately regulates the expression of EMT markers in the nucleus, promoting the metastasis of PCa (Figure 7G). These experiments confirmed that the activation of the TGFβ1/SMAD2/3 signaling pathway mediated by MMP9 is a potential mechanism for promoting the progression of PCa.
Discussion
In our study, we innovatively propose that high expression of SPP1 in M2 macrophages promotes the malignant progression of castration-resistant PCa. Mechanistically, we found that the upregulation of SPP1 in M2 macrophages increased the expression of MMP9 through activation of the PI3K/AKT signaling pathway. The increased secretion of MMP9 in M2 macrophages facilitated the TGFβ signaling pathway in PCa cells, thereby enhancing their invasive, migratory, and EMT transformation capabilities. Finally, we applied an SPP1 inhibitor in an orthotopic PCa metastasis model and observed a significant inhibition of tumor progression, along with a noticeable reduction in the recruitment of M2 macrophages in the TME.
PCa exhibits significant heterogeneity compared to other tumors, as the cold reactivity of immunotherapy distinguishes it significantly from other tumors (8). The TME is the main driving force for tumor occurrence and development, and its complex cellular components and intercellular communication are important factors that constitute this network. According to relevant research, TME plays an important role in tumor progression, metastasis, immune suppression, and drug resistance (21,22). Macrophages are representative cell types in the TME (10), and in recent years, SPP1 macrophages have been found to promote the progression of various tumors. In colon cancer, small cell lung cancer, and liver cancer, blocking the expression of SPP1 in macrophages has been found to increase sensitivity to immune therapy, thereby inhibiting tumor progression (23,24). However, it has not yet been confirmed whether SPP1 macrophages play a role in promoting castration-resistant PCa progression. Previous studies on the mechanisms of CRPC progression have primarily focused on SPP1+ cancer-associated fibroblast (CAF) cells, suggesting their high recruitment in the TME and close association with PCa progression (25,26). Nevertheless, our analysis of PCa single-cell data and mCRPC datasets revealed that M2 macrophages constitute the highest proportion in the metastatic microenvironment of CRPC. Additionally, there are currently related studies confirming the ability of M2 macrophages to promote CRPC progression (3,27). Notably, the expression of SPP1 in macrophages is also an indicator for evaluating macrophage polarization (14). When macrophages have high expression of SPP1, they polarize towards M2, which has also been confirmed in our experiment. There has been speculation regarding whether SPP1 macrophages promote the progression of CRPC. In the experiments we designed, we obtained verification that SPP1+ macrophages significantly enhance the invasion and migration of CPRC cells. Moreover, in in vivo experiments, the application of an SPP1 inhibitor effectively inhibited the progression of PCa and reduces the presence of M2 macrophages in the TME.
Interestingly, analysis of SPP1-related genes in PCa single-cell datasets and the SU2C-mCRPC cohort revealed that SPP1 is most significantly correlated with MMP9. Moreover, the PCa single-cell database showed that MMP9 is only highly expressed in macrophages, suggesting a potential regulatory relationship between SPP1 and MMP9 in M2 macrophages. Although previous research has confirmed that SPP1 can promote MMP9 expression in fibroblasts by activating the Akt signaling pathway (20), this regulatory mechanism has not been reported in PCa. Therefore, we confirmed in M2 macrophages that SPP1 regulates MMP9 expression through the PI3K/AKT signaling pathway and affects the malignant phenotype of PCa cells. MMP9, a member of the MMP family, has the function of cleaving LAP to activate TGF-β (20), which is consistent with the results of pathway enrichment and suggests that in M2 macrophages, increased MMP9 expression can activate the function of TGFβ1, leading to increased synthesis and secretion of mature TGFβ1. Upon binding to PCa cell receptors, the classic Smad2/3 signaling pathway is activated, regulating the expression of EMT-related molecules, and ultimately promoting PCa metastasis.
Conclusions
We have proposed for the first time that high expression of SPP1 in M2 macrophages is a key factor in the progression of CRPC. Through in-depth mechanistic analysis, we have confirmed the involvement of the SPP1-PI3K/AKT-MMP9 signaling axis. Furthermore, our further research has uncovered the MMP9-TGFβ1-Smad2/3 signaling axis. Connecting these signaling pathways provides a preliminary explanation for the potential mechanism by which high expression of SPP1 promotes CRPC metastasis in M2 macrophages. Therefore, targeting SPP1 and its associated signaling pathways holds great promise as a therapeutic approach for CRPC. It has significant clinical implications for inhibiting CRPC metastasis and prolonging patient survival.
Acknowledgments
Funding: This research was supported by
Footnote
Reporting Checklist: The authors have completed the MDAR and ARRIVE reporting checklists. Available at https://tau.amegroups.com/article/view/10.21037/tau-24-127/rc
Data Sharing Statement: Available at https://tau.amegroups.com/article/view/10.21037/tau-24-127/dss
Peer Review File: Available at https://tau.amegroups.com/article/view/10.21037/tau-24-127/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tau.amegroups.com/article/view/10.21037/tau-24-127/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). All animal studies carried out in this project strictly followed the regulations of the China Nature Conservation Committee and were approved by the Animal Experiment Welfare and Ethics Committee of the Army Medical University (No. AMUWEC20232943), in compliance with institutional guidelines for the care and use of animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Wagle NS, et al. Cancer statistics, 2023. CA Cancer J Clin 2023;73:17-48. [Crossref] [PubMed]
- Bishr M, Saad F. Overview of the latest treatments for castration-resistant prostate cancer. Nat Rev Urol 2013;10:522-8. [Crossref] [PubMed]
- Li XF, Selli C, Zhou HL, et al. Macrophages promote anti-androgen resistance in prostate cancer bone disease. J Exp Med 2023;220:e20221007. [Crossref] [PubMed]
- Chhabra Y, Weeraratna AT. Fibroblasts in cancer: Unity in heterogeneity. Cell 2023;186:1580-609. [Crossref] [PubMed]
- Caronni N, La Terza F, Vittoria FM, et al. IL-1β(+) macrophages fuel pathogenic inflammation in pancreatic cancer. Nature 2023;623:415-22. [Crossref] [PubMed]
- Hofbauer LC, Bozec A, Rauner M, et al. Novel approaches to target the microenvironment of bone metastasis. Nat Rev Clin Oncol 2021;18:488-505. [Crossref] [PubMed]
- Sternberg C, Armstrong A, Pili R, et al. Randomized, Double-Blind, Placebo-Controlled Phase III Study of Tasquinimod in Men With Metastatic Castration-Resistant Prostate Cancer. J Clin Oncol 2016;34:2636-43. [Crossref] [PubMed]
- Tong D. Selective estrogen receptor modulators contribute to prostate cancer treatment by regulating the tumor immune microenvironment. J Immunother Cancer 2022;10:e002944. [Crossref] [PubMed]
- Wang D, Cheng C, Chen X, et al. IL-1β Is an Androgen-Responsive Target in Macrophages for Immunotherapy of Prostate Cancer. Adv Sci (Weinh) 2023;10:e2206889. [Crossref] [PubMed]
- Komohara Y, Fujiwara Y, Ohnishi K, et al. Tumor-associated macrophages: Potential therapeutic targets for anti-cancer therapy. Adv Drug Deliv Rev 2016;99:180-5. [Crossref] [PubMed]
- Matsubara E, Yano H, Pan C, et al. The Significance of SPP1 in Lung Cancers and Its Impact as a Marker for Protumor Tumor-Associated Macrophages. Cancers (Basel) 2023;15:2250. [Crossref] [PubMed]
- Inoue M, Shinohara ML. Intracellular osteopontin (iOPN) and immunity. Immunol Res 2011;49:160-72. [Crossref] [PubMed]
- Zhao H, Chen Q, Alam A, et al. The role of osteopontin in the progression of solid organ tumour. Cell Death Dis 2018;9:356. [Crossref] [PubMed]
- Bill R, Wirapati P, Messemaker M, et al. CXCL9:SPP1 macrophage polarity identifies a network of cellular programs that control human cancers. Science 2023;381:515-24. [Crossref] [PubMed]
- Sathe A, Mason K, Grimes SM, et al. Colorectal Cancer Metastases in the Liver Establish Immunosuppressive Spatial Networking between Tumor-Associated SPP1+ Macrophages and Fibroblasts. Clin Cancer Res 2023;29:244-60. [Crossref] [PubMed]
- Liu Y, Zhang Q, Xing B, et al. Immune phenotypic linkage between colorectal cancer and liver metastasis. Cancer Cell 2022;40:424-437.e5. [Crossref] [PubMed]
- Qi J, Sun H, Zhang Y, et al. Single-cell and spatial analysis reveal interaction of FAP(+) fibroblasts and SPP1(+) macrophages in colorectal cancer. Nat Commun 2022;13:1742. [Crossref] [PubMed]
- Ramadan A, Afifi N, Yassin NZ, et al. Mesalazine, an osteopontin inhibitor: The potential prophylactic and remedial roles in induced liver fibrosis in rats. Chem Biol Interact 2018;289:109-18. [Crossref] [PubMed]
- Massam-Wu T, Chiu M, Choudhury R, et al. Assembly of fibrillin microfibrils governs extracellular deposition of latent TGF beta. J Cell Sci 2010;123:3006-18. [Crossref] [PubMed]
- Kramerova I, Kumagai-Cresse C, Ermolova N, et al. Spp1 (osteopontin) promotes TGFβ processing in fibroblasts of dystrophin-deficient muscles through matrix metalloproteinases. Hum Mol Genet 2019;28:3431-42. [Crossref] [PubMed]
- Wei R, Liu S, Zhang S, et al. Cellular and Extracellular Components in Tumor Microenvironment and Their Application in Early Diagnosis of Cancers. Anal Cell Pathol (Amst) 2020;2020:6283796. [Crossref] [PubMed]
- Quail DF, Joyce JA. Microenvironmental regulation of tumor progression and metastasis. Nat Med 2013;19:1423-37. [Crossref] [PubMed]
- Liu Y, Xun Z, Ma K, et al. Identification of a tumour immune barrier in the HCC microenvironment that determines the efficacy of immunotherapy. J Hepatol 2023;78:770-82. [Crossref] [PubMed]
- Hu J, Zhang L, Xia H, et al. Tumor microenvironment remodeling after neoadjuvant immunotherapy in non-small cell lung cancer revealed by single-cell RNA sequencing. Genome Med 2023;15:14. [Crossref] [PubMed]
- Wang H, Li N, Liu Q, et al. Antiandrogen treatment induces stromal cell reprogramming to promote castration resistance in prostate cancer. Cancer Cell 2023;41:1345-1362.e9. [Crossref] [PubMed]
- Li X, Mu P. The Critical Interplay of CAF Plasticity and Resistance in Prostate Cancer. Cancer Res 2023;83:2990-2. [Crossref] [PubMed]
- Salachan PV, Rasmussen M, Ulhøi BP, et al. Spatial whole transcriptome profiling of primary tumor from patients with metastatic prostate cancer. Int J Cancer 2023;153:2055-67. [Crossref] [PubMed]
(English Language Editor: J. Jones)


