
LncRNA ZFHX4-AS1 as a novel biomarker in adrenocortical carcinoma
Highlight box
Key findings
• This study demonstrates that the long non-coding RNA (lncRNA) ZFHX4 antisense RNA 1 (ZFHX4-AS1) can serve as a new biomarker for adrenocortical carcinoma (ACC) and validates its biological function through in vitro experiments.
What is known and what is new?
• ZFHX4-AS1 is a recently discovered lncRNA situated on chromosome 8q21.13 and is closely associated with chromosome 8q21.11 deletion syndrome. ZFHX4-AS1 is upregulated in various types of cancer and plays a significant role in cancer growth and metastasis. However, the prognosis and regulatory mechanism of ZFHX4-AS1 in ACC are still unclear.
• In this study, we integrated cohort data from multiple centers and utilized bioinformatics analysis along with in vitro experiments to investigate the correlation between ZFHX4-AS1 and the prognosis of ACC patients, as well as the biological function of ZFHX4-AS1.
What is the implication, and what should change now?
• The results indicate that ZFHX4-AS1 can accurately predict the prognosis of ACC patients and aid in customizing risk stratification for ACC patients. In the future, we hope to gather additional clinical samples for verification.
Introduction
Adrenocortical carcinoma (ACC) is classified as an uncommon and malignant endocrine neoplasm. According to statistics, the annual incidence rate of ACC in adults is 0.5 to 2 cases per million people (1,2). The annual incidence rate in children worldwide is estimated to be 0.2 to 0.3 cases per million children, while in southern Brazil, the incidence rate for pediatric ACC is significantly higher, ranging from 2.9 to 4.2 cases per million children (3). ACC often presents with insidious early symptoms, and many patients are diagnosed when the tumor has already invaded locally or metastasized, leading to a poor prognosis, resulting in overall survival (OS) rate of 37–59% over a 5-year period (4,5). Currently, radical surgery remains the only effective cure for localized ACC (6). However, even after complete tumor resection, up to 25% of ACC patients experience local recurrence or distant metastasis (7). For these patients who are at a high risk of recurrence or metastasis, more aggressive and reliable clinical treatment strategies are adopted to improve prognosis, including chemotherapy, radiotherapy, and targeted therapy (1). Precise risk stratification, and prognosis prediction for ACC patients remain key issues that urgently need to be addressed in clinical diagnosis and treatment.
In recent years, clinical indicators such as the tumor proliferation index (Ki-67), tumor stage, the status of surgical margins, and hormone production capacity (CYP11A1, CYP17A1) have been widely used to predict the prognosis of ACC patient (8-10). In addition, some biomarkers, such as BUB1B-PINK1, IGF2 and GOS2, are also used to predict the diagnosis and prognosis of patients with ACC (11-14). However, due to the impact of tumor heterogeneity (15), these clinical indicators are gradually unable to meet the needs of individualized diagnosis and prognosis assessment for ACC patients (16). As technology like high-throughput sequencing develops so quickly, long non-coding RNA (lncRNA), known for their complex and precise regulatory functions, have gradually gained attention in the fields of biology and medicine (17). A substantial body of research has revealed the indispensable role of lncRNA in various biological processes of human malignancies, including initiation, progression, exacerbation, and metastasis (18,19). A large number of studies have shown that, in addition to BUB1B-PINK1, IGF2, and GOS2, a significant number of miRNAs and lncRNAs are also utilized in the diagnosis and prognosis prediction of ACC (20). In ACC, miR-100 can activate the MAPK signaling pathway through CXCR7 to induce tumor progression, and some patients currently benefit from CXCR7-targeted therapy (21). In vitro studies have indicated that miR-375 plays a role in the pathogenesis of ACC by regulating the PI3K/Akt pathway (22). Circ-CCAC1 overexpressed in ACC can enhance ACC cell proliferation, migration, and invasion by modulating miR-514a-5p, and is linked to unfavorable patient prognosis (23). Therefore, lncRNA not only serve as biomarkers for tumors but also have potential as targets for clinical therapy, making their role in cancer worthy of further investigation (24).
Recent findings indicate that lncRNA have intricate and specific regulatory functions in the development and advancement of cancer, primarily through their roles as tumor suppressors or oncogenes (25). A few lncRNAs contribute to the development of cancer by acting as oncogenic agents. They may promote the growth and spread of cancer cells, accelerating the progression of the disease (23). By interacting with the translocated promoter region (TPR), TLNC1 can induce TPR-mediated p53 trafficking from the nucleus to the cytoplasm, which subsequently suppresses the transcription of p53 target genes and ultimately contributes to the development of liver cancer (26). Additionally, the BREA2 plays a critical role in the progression and metastasis of breast cancer. The gene is significantly upregulated in advanced breast cancer and is associated with a poor patient prognosis (27). It functions by impeding the proteasome-mediated degradation of the Notch1 intracellular domain (NICD1) through its interaction with the WW domain-containing E3 ubiquitin protein ligase 2 (WWP2) (28). Numerous studies suggest that H19 and the well-known lncRNA HOX transcript antisense intergenic RNA (HOTAIR) act as oncogenes, and their expression is strongly correlated with the stage and prognosis of various malignancies (29-31). Certain lncRNA have also demonstrated the ability to concurrently impede the progression and dissemination of cancer cells. It has tumor-suppressive properties. For instance, Zhang et al. discovered that there is a decrease in the expression levels of lncRNA BC069792. BC069792 acts as a molecular sponge, adsorbing hsa-miR-658 and hsa-miR-4739, thereby upregulating the protein expression of Potassium Voltage-Gated Channel Q4 (KCNQ4) and inhibiting the growth of breast cancer (32). It can be used as a novel diagnostic marker and therapeutic target for breast cancer. One potential novel treatment target and diagnostic indicator for breast cancer is with the mechanism of cancer growth (32). Furthermore, by targeting the LENGA/BRD7/TP53 axis, the lncRNA LENGA inhibits the proliferation and migration of gastric cancer cells. In summary, these findings reveal the complex roles of lncRNA in cancer pathogenesis and provide new opportunities for cancer detection and treatment (33). Nonetheless, further research is needed to fully understand the role that lncRNA play in cancer formation.
LncRNA ZFHX4 antisense RNA 1 (ZFHX4-AS1) was initially identified in association with the 8q21.11 deletion syndrome and exhibits abnormal expression in bladder, breast, and ovarian cancer tissues, serving as a biomarker indicative of poor prognosis in patients (34-37). Despite its significant impact on the growth and metastasis of various tumors, research on ZFHX4-AS1 in ACC has not been reported to date. As illustrated in Figure 1, our study employed a workflow that involved downloading data from four ACC patient cohorts from The Cancer Genome Atlas (TCGA) program and the Gene Expression Omnibus (GEO). We discovered that ZFHX4-AS1 holds substantial predictive power for the prognosis of ACC patients. We further explored the role of ZFHX4-AS1 in ACC through various approaches including gene set enrichment analysis (GSEA), mutation landscape analysis, and competing endogenous RNA (ceRNA) network analysis. Additionally, the oncogenic effects of ZFHX4-AS1 on ACC cell lines were validated through in vitro experiments. In conclusion, ZFHX4-AS1 could serve as a prognostic biomarker that can help with ACC risk stratification, according to our research findings. We present this article in accordance with the TRIPOD reporting checklist (available at https://tau.amegroups.com/article/view/10.21037/tau-23-649/rc).
Methods
Processing and access on public data
We downloaded RNA-Seq, clinical information, single nucleotide polymorphism (SNP), and copy number variation (CNV) for TCGA-ACC from the UCSC Xena website (https://xena.ucsc.edu/). Additionally, transcriptome data and clinical information for GSE10927 (11), GSE19750 (38), and GSE76019 (39) datasets were obtained from the GEO database (https://www.ncbi.nlm.nih.gov/geo/). We then excluded samples from these cohorts with OS, progression-free interval (PFI), and event-free survival (EFS) of less than 30 days. Finally, we utilized survminer package to determine the optimal cutoff value and categorize ACC patients into ZFHX4-AS1 high-expression and low-expression groups. Table S1 summarizes the detailed clinical characteristics of each cohort. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
GSEA
Gene sets “c2.cp.kegg.v7.4.entrez.gmt” and “c5.go.v7.4.entrez.gmt” were downloaded from the Molecular Signatures Database (MSigDB) (https://www.gsea-msigdb.org/gsea/msigdb) (40). We employed Spearman correlation analysis to assess the correlation between ZFHX4-AS1 and all mRNAs (41). All mRNAs were then ranked in descending order based on correlation coefficients. The clusterProfiler package was used to perform GSEA on the ranked list of mRNAs to identify significantly enriched functional pathways (42). False discovery rate (FDR) <0.001 was considered significant enrichment.
The landscape of somatic mutation
Mutation Annotation Format (MAF) files obtained from the UCSC Xena website were analyzed using the maftools package. To determine the mutation status of genes associated with ACC in both the high and low ZFHX4-AS1 expression groups, waterfall plots were generated to analyze the top 20 genes with somatic SNP mutations. CNV data for TCGA-ACC was downloaded from the UCSC Xena website and analyzed online using the GISTIC_2.0 module (https://www.genepattern.org/modules/docs/GISTIC_2.0) on the GenePattern website to identify regions with CNV in both ZFHX4-AS1 expression groups. Finally, the differences between the two groups’ CNV and gene mutation frequency between the two groups were compared using the Wilcoxon rank-sum test (43).
Cell culture
The ACC cell lines H295R and SW-13 were both acquired from Procell Biotechnology (Wuhan, China). H295R cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM)/F12 medium containing insulin-transferrin-selenium (ITS-G), 10% fetal bovine serum (FBS), and 1% penicillin and streptomycin. SW-13 cells were maintained in DMEM medium supplemented with 10% FBS and 1% penicillin and streptomycin. All the cell lines were cultured at 37 ℃ with 5% CO2 in a humidified incubator.
RNA interference and loss of function assays
Small interfering RNAs (siRNAs) targeting lncRNA ZFHX4-AS1 (si-ZFHX4-AS1#1: UAUGAUAGACAUUUCUACC; si-ZFHX4-AS1#2: UAGUAAUGUUGUUACAUCC; si-ZFHX4-AS1#3: UGUAACAUUAUGAAUCUGG) were synthesized by Genepharma (Suzhou, China). The RNAi-Mate reagent provided by Genepharma was used for stable transfection, and the cells were co-cultured for 72 hours after forming a complex with siRNA. The transfection effectiveness of siRNAs was confirmed using quantitative reverse transcription polymerase chain reaction (qRT-PCR). The proliferative capabilities of ACC cell lines post-ZFHX4-AS1 knockdown were assessed using the Cell Counting Kit-8 (CCK-8) and colony formation assays. Transwell assays was employed to evaluate the migratory ability of cells with ZFHX4-AS1 knockdown. For CCK-8 and Transwell assays, cells were gently fixed with 4% paraformaldehyde for 10 minutes and then stained with 0.1% crystal violet for 30 minutes. After thorough cleaning and drying, observations were made using an inverted microscope, and statistical calculations were conducted. All experiments were performed in triplicate.
OE_ZFHX4-AS1 construction and treatment
Using the pCDH-CMV-MCS-EF1-CopGFP-T2A-Puro vector labeled with green fluorescent protein (GFP), a model for overexpression of the ZFHX4-AS1 gene was constructed. The pCDH-CMV-MCS-EF1-CopGFP-T2A-Puro-ZFHX4-AS1_shRNA (5'-GCAGGCGCCACAGAGAATAG-3') vector was provided by Guangzhou Elite Bio-Tech Co., Ltd. (Guangzhou, China). Lipofectamine 2000, provided by Invitrogen (California, USA), was used to transfect H295R cells following the manufacturer’s instructions, resulting in the establishment of stable ZFHX4-AS1 overexpression (OE_ZFHX4-AS1) cells. H295R cells transfected with the empty pCDH-CMV-MCS-EF1-CopGFP-T2A-Puro vector (OE_NC) were used as controls. The transfection rate was evaluated after 48 hours. For CCK-8 and Transwell assays, cells were gently fixed with 4 paraformaldehyde for 10 minutes and then stained with 0.1% crystal violet for 30 minutes. All experiments were performed in triplicate.
Statistical analysis
Data analysis and visualization were conducted using R software (v4.1.0, https://www.r-project.org/) and Graphpad Prism 8. Spearman correlation analysis was used to compute correlation coefficients. The Wilcoxon rank-sum test or Student’s t-tests was employed for comparing differences between two continuous variables. The survminer and survival packages were utilized to determine the optimal cutoff values and conduct Kaplan-Meier survival analysis. The timeROC package was utilized to calculate the area under the curve (AUC) value for the time-dependent receiver operating characteristic (ROC) curve, and nomograms were constructed using the rms package. All analyses employed a two-tailed design, with P value of less than 0.05 deemed statistically significant.
Results
Prognostic value of ZFHX4-AS1
Based on multi-center ACC cohorts from TCGA and GEO, we identified ZFHX4-AS1 as a potential biomarker. We performed a Kaplan-Meier survival analysis after dividing the expression levels of ZFHX4-AS1 into the high and low expression groups in order to find out the correlation between ZFHX4-AS1 expression and the prognosis of ACC patients. In the TCGA-ACC cohort, patients who had high ZFHX4-AS1 expression had worse OS and PFI (P=0.02, P<0.001) (Figure 2A,2B). Similarly, in the GSE10927 and GSE19750 cohorts, patients with low ZFHX4-AS1 expression group had more satisfactory OS compared to those with high expression (P=0.02, P=0.02) (Figure 2C,2D). Additionally, in the pediatric cohort GSE76019, a higher expression of ZFHX4-AS1 was linked to a lower EFS (P=0.001) (Figure 2E). ROC curve results showed that in TCGA-ACC, the AUC values for 1-, 3- and 5-year OS were 0.970, 0.585, and 0.716, correspondingly, and for 1-, 3-, and 5-year PFI were 0.713, 0.655 and 0.764. In GSE10927, the AUC values for 1-, 3- and 5-year OS were 0.859, 0.610 and 0.603; in GSE19750, they were 0.729, 0.611 and 0.628; in GSE76019, the AUC values for 1-, 3- and 5-year EFS were 0.792, 0.778 and 0.925 (Figure 2F). According to the results, ZFHX4-AS1 provides a strong predictive performance for the prognosis of ACC patients. Cox regression analysis, both univariate and multivariate, showed that in the TCGA-ACC, GSE10927 and GSE19750 cohorts, ZFHX4-AS1 was strongly linked to low OS and PFI in ACC patients [P<0.05, hazard ratio (HR) >1.5] (Table 1). Univariate Cox regression analysis in the GSE76019 cohort showed that EFS in ACC patients was closely related to ZFHX4-AS1, gender, age and stage (P<0.01, HR >1).
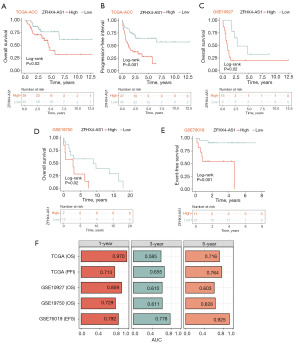
Table 1
| Characteristics | Univariate analysis | Multivariate analysis | |||
|---|---|---|---|---|---|
| HR (95% CI) | P value | HR (95% CI) | P value | ||
| TCGA-ACC (OS) | |||||
| ZFHX4-AS1 | 1.753 (1.143–2.689) | 0.01 | 1.606 (1.032–2.499) | 0.04 | |
| Age | 1.014 (0.990–1.039) | 0.26 | |||
| Gender | 0.205 (0.089–0.470) | <0.001 | 0.223 (0.096–0.516) | <0.001 | |
| Stage | 1.103 (0.732–1.663) | 0.64 | |||
| pT | 1.218 (0.817–1.816) | 0.33 | |||
| pN | 0.576 (0.171–1.941) | 0.37 | |||
| TCGA-ACC (PFI) | |||||
| ZFHX4-AS1 | 2.125 (1.478–3.056) | <0.001 | 2.095 (1.449–3.029) | <0.001 | |
| Age | 1.003 (0.983–1.024) | 0.77 | |||
| Gender | 0.382 (0.203–0.721) | 0.003 | 0.403 (0.213–0.761) | 0.005 | |
| Stage | 1.243 (0.888–1.740) | 0.21 | |||
| pT | 1.232 (0.889–1.706) | 0.21 | |||
| pN | 0.941 (0.392–2.262) | 0.89 | |||
| GSE10927 | |||||
| ZFHX4-AS1 | 11.371 (2.523–51.249) | 0.002 | 11.861 (2.424–58.028) | 0.002 | |
| Age | 1.011 (0.964–1.061) | 0.64 | |||
| Gender | 2.804 (1.030–7.638) | 0.04 | 2.742 (0.982–7.655) | 0.054 | |
| GSE19750 | |||||
| ZFHX4-AS1 | 2.067 (1.215–3.517) | 0.007 | 1.887 (1.072–3.323) | 0.03 | |
| Age | 1.035 (0.994–1.079) | 0.10 | 1.023 (0.980–1.068) | 0.30 | |
| Gender | 1.262 (0.463–3.441) | 0.65 | |||
| Stage | 1.027 (0.739–1.428) | 0.87 | |||
| GSE76019 | |||||
| ZFHX4-AS1 | 1.901 (1.223–2.954) | 0.004 | 1.522 (0.837–2.765) | 0.17 | |
| Age | 1.019 (1.010–1.029) | <0.001 | 1.014 (1.000–1.028) | 0.057 | |
| Stage | 3.075 (1.468–6.442) | 0.003 | 1.452 (0.548–3.847) | 0.45 | |
| Gender | 5.684 (1.697–19.003) | 0.005 | 3.717 (0.937–14.749) | 0.06 | |
HR, hazard ratio; CI, confidence interval; TCGA, The Cancer Genome Atlas; ACC, adrenocortical carcinoma; OS, overall survival; PFI, progression-free interval.
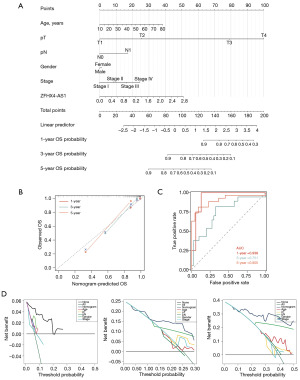
Recently, nomograms have been widely utilized as risk assessment tools for prognostication in cancer patients. To enhance the clinical applicability of this study, we constructed nomograms based on scores of predictive variables, including ZFHX4-AS1, age, T stage, N stage and gender to predict the 1-, 3- and 5-year OS and PFI for ACC patients (Figure 3A, Figure S1A). Calibration curves demonstrated that the estimated 1-, 3- and 5-year OS and PFI by the nomograms closely matched the actual outcomes (Figure 3B, Figure S1B). Moreover, the AUC values for 1-, 3- and 5-year OS of the nomograms were 0.939, 0.751 and 0.905, respectively, and those for 1-, 3- and 5-year PFI were 0.744, 0.763 and 0.855 (Figure 3C, Figure S1C), indicating that our constructed nomograms are accurate and reliable. Decision curve analysis (DCA), which estimates the net benefit of a model according to the variation in the number of true positives and false positives, is frequently used to assess the effectiveness of nomograms in aiding clinical decisions. The DCA results suggest that using nomograms for predicting the 1-, 3- and 5-year survival rates of ACC patients offers a greater net benefit and is more effective than other clinical indicators (Figure 3D, Figure S1D). In summary, ZFHX4-AS1 is a reliable novel biomarker that can effectively forecast for ACC patient’s prognosis.
GSEA
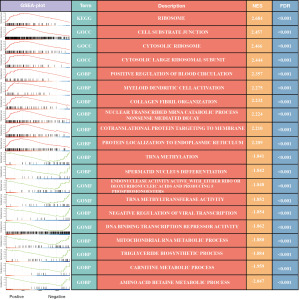
Additionally, we conducted a GSEA to probe into the potential biological functions of ZFHX4-AS1 in ACC (available online: https://cdn.amegroups.cn/static/public/tau-23-649-1.xlsx). We selected the top 10 positively and negatively correlated pathways based on the Normalized Enrichment Score (NES), and FDR <0.001. As shown in Figure 4, the high expression group of ZFHX4-AS1 predominantly exhibited enrichment in pathways associated with protein synthesis and translation. These include ribosome, cytoplasmic ribosomes, protein localization to the endoplasmic reticulum, and cotranslational protein targeting to the membrane. On the other hand, the low expression group of ZFHX4-AS1 was primarily linked to metabolic pathways, such as amino acid betaine metabolism, carnitine metabolism, triglyceride biosynthesis, and mitochondrial RNA metabolism.
ZFHX4-AS1 promotes proliferation and migration in ACC
The majority of the above analyses were in silico. Therefore in vitro experimental validation was conducted to assess the role of ZFXH4-AS1 in ACC. Initially, specific primers targeting ZFHX4-AS1 were designed, and their transfection efficiency was verified using qRT-PCR. Compared to the control group si-NC, all three primers significantly downregulated the expression of ZFHX4-AS1 in ACC cell lines (H295R and SW-13), with si-ZFHX4-AS1#1 and si-ZFHX4-AS1#3 showing higher efficiency (P=0.01) (Figure 5A). Subsequently, The CCK-8 assay was used to evaluate the ZFHX4-AS1 knockdown cells’ capacity for proliferation. Over a period of five days, knockdown of ZFHX4-AS1 notably slowed the proliferation of ACC cells (P<0.001) (Figure 5B). Additionally, the impact of ZFHX4-AS1 knockdown on cell proliferation was studied using a colony formation assay. As shown in Figure 5C, compared to the control group, cells with ZFHX4-AS1 knockdown exhibited reduced viability (P=0.001, P<0.001). Transwell assays confirmed that the migratory ability of ACC cells was diminished following the downregulation of ZFHX4-AS1 (P=0.04, P<0.001) (Figure 5D). Overall, our preliminary findings confirm that ZFHX4-AS1 plays a promotive role in the proliferation, and migration abilities of ACC.
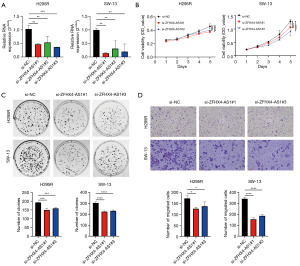
OE_ZFHX4-AS1 promotes ACC cell proliferation and migration
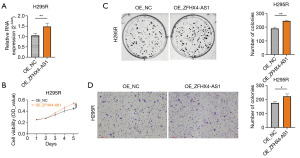
Through in vitro cell experiments involving the knockdown of ZFHX4-AS1 (pCDH-CMV-MCS-EF1-CopGFP-T2A-Puro-ZFHX4-AS1_shRNA), we observed that elevated ZFHX4-AS1 expression can enhance the proliferation and invasion of ACC cells. To further support this finding, we designed specific vectors and verified their transfection efficiency using qRT-PCR. Compared with the control group OE-NC, OE_ZFHX4-AS1 significantly increased the expression of ZFHX4-AS1 in H295R (P=0.01) (Figure 6A). Similarly, CCK-8 experiments showed that overexpression of ZFHX4-AS1 significantly accelerated the proliferation of H295R cells within five days (P<0.001) (Figure 6B). Colony formation experiments suggest that cells overexpressing ZFHX4-AS1 exhibited increased proliferation compared to OE_NC (P=0.01) (Figure 6C). In addition, the Transwell assay confirmed that the migration ability of H295R cells was significantly enhanced after overexpression of ZFHX4-AS1 (P=0.04) (Figure 6D). Overall, we demonstrated through knockdown and overexpression experiments that ZFHX4-AS1 plays a promoting role in the proliferation and migration capabilities of ACC.
Construction of the ceRNA network
Our in vitro experiments demonstrated that ZFHX4-AS1 contributes to the development and progression of ACC, yet the regulatory mechanisms of ZFHX4-AS1 in ACC are not fully understood. In this study, we utilized the “GDCRNATools” package within the TCGA-ACC dataset to explore the potential lncRNA-miRNA-mRNA ceRNA regulatory network. Adhering to our inclusion criteria (P<0.05 and |correlation coefficient| >0.4), we identified a ceRNA network comprising 70 molecules, which includes 1 lncRNA, 28 mRNAs and 41 miRNAs. This network was visualized using Cytoscape (Version 3.1.0). As depicted in Figure 7, the central red square region represents the lncRNA ZFHX4-AS1, the middle orange triangular area signifies the 28 mRNAs, and the outer green circular region denotes the 41 miRNAs.
Somatic mutation landscape of ZFHX4-AS1
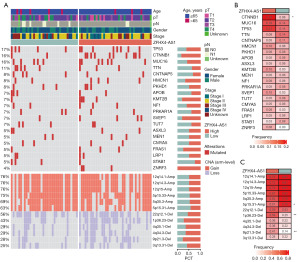
To gain deeper insight into the mutational landscape of ZFHX4-AS1, we identified 20 frequently mutated genes, including TP53, CTNNB1, MUC16, TTN, CNTNAP5, HMCN1, PKHD1, APOB, KMT2B, NF1, PRKAR1A, SVEP1, TUT7, ASXL3, MEN1, CMYA5, FRAS1, LRP1, STAB1 and ZNRF3. We also examined the CNV status of the top six amplified (Amp) and homozygously deleted (Del) chromosomal segments between the ZFHX4-AS1 high and low expression groups (Figure 8A). Notably, the mutation frequency of CTNNB1 (P=0.02) showed a vital difference between the ZFHX4-AS1 high and low expression groups (Figure 8B). Furthermore, we investigated the CNV status of Amp and Del chromosomal segments between the ZFHX4-AS1 high and low expression groups. As indicated in Figure 8C, in comparison to the high expression group of ZFHX4-AS1, the CNV frequencies of 5p15.31-Amp were considerably greater in the low expression group (P=0.04). In contrast, 1p36.23-Del and 9p21.3-Del exhibited a higher CNV frequency in the high expression group of ZFHX4-AS1 (P=0.01).
Discussion
ACC is a kind of cancer that arises from the adrenal cortex. It is characterized by its strong invasiveness, high recurrence rate, and propensity for metastasis (1). The rarity of ACC, coupled with its variable and complex clinical presentations, makes early diagnosis challenging (5,44). Statistics show that only about 10% of cases are incidentally discovered during physical examinations, with the majority of patients presenting at advanced stages (III or IV) when the optimal window for clinical intervention has passed (45). Currently, mitotane is the preferred adjuvant treatment drug recommended by multiple international guidelines and consensuses and is the only drug approved by the US and European regulatory authorities for treating ACC (7,46). However, mitotane can cause severe adverse reactions like neurotoxicity, liver impairment, and fractures, and its safety and efficacy remain controversial. Prospective data indicate that the 5-year OS for stage II ACC patients after complete tumor resection can be as high as 90%, whereas it is only about 15% for patients with advanced ACC (47). Identifying novel biomarkers is critical for ACC patients’ early diagnosis and prognosis prediction, and it can help to better tailor therapy management.
LncRNA are non-protein-coding RNAs with transcript lengths exceeding 200 nucleotides (48). LncRNA are classified into five categories based on their chromosomal position: intronic lncRNA, intergenic lncRNA, sense lncRNA, antisense lncRNA, and bidirectional lncRNA (49). Antisense lncRNA, constituting approximately 40% of total lncRNA, are located near coding genes but transcribed in the opposite direction. Functionally, antisense lncRNA play multiple roles in various biological processes, including embryonic pluripotency, differentiation, and development, and are closely associated with disease progression (50). Numerous studies have reported that dysregulation of AS-lncRNA often promotes tumor development by enhancing the proliferative, invasive, and metastatic capabilities of tumor cells (19,51). ZFHX4 is one of the five members of the zinc finger homeobox family, with a molecular weight of 397 kDa. This protein contains four homologous domains and 22 zinc finger structures, located at 8q13.3-q21.1 (52). ZFHX4-AS1 is a newly discovered lncRNA that targets the antisense sequence of the ZFHX4 gene, located on chromosome 8q21.13 (34). There are few reports on the functional study of ZFHX4-AS1 in tumors. ZFHX4-AS1 was initially identified as significantly overexpressed in breast cancer, promoting cell apoptosis by inhibiting the Hippo signaling pathway (36). Additionally, it is currently recognized as a prognostic marker for ovarian cancer, breast cancer, and bladder cancer, predicting the prognosis of tumor patients. Studies have shown its association with thyroid-associated ophthalmopathy, gastric cancer, and susceptibility to glaucoma. Furthermore, it can function as a ceRNA to influence the severity of schizophrenia (53-55). This RNA acts as an oncogene in bladder cancer and breast cancer, but its role in ACC is not yet clear.
In recent years, the growing significance of lncRNA in cancer has become increasingly prominent (24). Therefore, it is crucial to study the upstream regulatory factors and downstream target genes associated with lncRNA to gain a comprehensive understanding of cancer progression and to identify potential therapeutic targets. There is ample evidence that lncRNA can function as a scaffold, decoy, ceRNA and promote DNA damage, immune evasion, and cellular metabolic disorders in cancer cells through post-translational modifications of key proteins, including ubiquitination, phosphorylation, and acetylation (18,25,56,57). Among these, regulation through ceRNA is widely considered a major mechanism of lncRNA regulation (58). Through the regulation of ceRNA, lncRNA can compete with miRNA for binding, thereby influencing the expression of miRNA’s target genes and regulating various biological processes in cells. This regulatory mechanism is complex and delicate, necessitating precise control and equilibrium to ensure the normal advancement of various biological processes in organisms (59). For instance, the lncRNA LINC00680 functions as a ceRNA, facilitating the invasion and advancement of esophageal squamous cell carcinoma through the miR-423-5p/PAK6 axis. Lin et al.’s study demonstrated that ITGB8-AS1 functions as a ceRNA to control the proliferation of colon cancer cells and tumor growth by regulating focal adhesion signaling (57). Research by Wang et al. demonstrates that ZFHX4-AS1 is significantly expressed in bladder cancer, and can promote the development and metastasis of bladder cancer through the ceRNA network (35). Therefore, we believe that ZFHX4-AS1 may also function in a similar mechanism in ACC.
In this study, we first analyzed the prognostic value of ZFHX4-AS1 expression levels in ACC patients using clinical cohorts from TCGA and GEO databases. Comprehensive analysis of OS, PFI and EFS in ACC cohorts revealed that patients with high ZFHX4-AS1 expression group had substantially worse overall prognosis compared to those with low expression. Furthermore, it was verified by univariate and multivariate cox regression analyses that ZFHX4-AS1 might function as a standalone risk factor for ACC patients’ prognosis. However, in the GSE76019 cohort, multivariate cox regression analysis indicated no significant correlation between ZFHX4-AS1 and EFS in ACC patients. This could be caused by inconsistent sequencing platforms or a tiny sample size. Subsequent in vitro experiments demonstrated that knockdown of ZFHX4-AS1 inhibited proliferation, clonogenicity, and migration abilities of ACC cell lines. These data imply that ZFHX4-AS1 is a reliable biomarker for predicting the prognosis of ACC patients.
Cancer, characterized by abnormal and uncontrolled cell proliferation, is primarily caused by genetic mutations (60). In this study, the most common mutated genes in ACC were TP53 and CTNNB1, consistent with previous research (61). Notably, CTNNB1 mutations were significantly more prevalent in the high ZFHX4-AS1 expression group. CTNNB1 encodes beta-catenin, a crucial component of the Wnt signaling pathway (62). Studies by Gaujoux et al. have shown that activation of the CTNNB1-mediated Wnt/β-catenin signaling pathway in ACC reduces patient OS and disease-free survival (63). Activation of this pathway has been implicated in tumor invasion, metastasis, and drug resistance in previous studies (64). and multiple research findings support the importance of Wnt/β-catenin signaling pathway activation in ACC (65,66). However, whether ZFHX4-AS1 affects ACC progression through CTNNB1 mutations requires further investigation. Additionally, the CNV frequency of 9p21.3-Del was significantly higher in the high ZFHX4-AS1 expression group. Barriga et al. noted that 9p21.3-Del is one of the most common homozygous deletions in human cancers, promoting tumor immune evasion, metastasis, and drug resistance (67,68), and is a contributing factor to the failure of immunotherapy in malignancies such as renal cell carcinoma, esophageal cancer, bladder cancer, and glioma (69,70). The deletion of human chromosome 9p21.3 is emerging as a biomarker indicating poor response to cancer therapy (71), warranting further investigation in ACC.
Despite the promising results indicating ZFHX4-AS1 as a biomarker, our study has limitations. Firstly, our data are mainly derived from retrospective samples; prospective multicenter samples are needed to validate the predictive value of ZFHX4-AS1. Secondly, incomplete clinical information (pathological staging, age, etc.) in some public datasets may have overlooked potential relationships between ZFHX4-AS1 and certain variables; more detailed clinical data on ACC patients are needed for further validation.
Conclusions
In this study, we found that lncRNA ZFHX4-AS1 accurately stratifies risk in ACC patients, demonstrating robust predictive capability, based on four multicenter cohorts from TCGA and GEO. Additionally, cell experiments further validated the oncogenic effects of ZFHX4-AS1 on ACC cell lines. In conclusion, ZFHX4-AS1 is a potential molecular marker indicating poor prognosis in ACC patients.
Acknowledgments
Funding: This work was supported by
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://tau.amegroups.com/article/view/10.21037/tau-23-649/rc
Data Sharing Statement: Available at https://tau.amegroups.com/article/view/10.21037/tau-23-649/dss
Peer Review File: Available at https://tau.amegroups.com/article/view/10.21037/tau-23-649/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tau.amegroups.com/article/view/10.21037/tau-23-649/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Else T, Kim AC, Sabolch A, et al. Adrenocortical carcinoma. Endocr Rev 2014;35:282-326. [Crossref] [PubMed]
- Sharma E, Dahal S, Sharma P, et al. The Characteristics and Trends in Adrenocortical Carcinoma: A United States Population Based Study. J Clin Med Res 2018;10:636-40. [Crossref] [PubMed]
- McAteer JP, Huaco JA, Gow KW. Predictors of survival in pediatric adrenocortical carcinoma: a Surveillance, Epidemiology, and End Results (SEER) program study. J Pediatr Surg 2013;48:1025-31. [Crossref] [PubMed]
- Jouinot A, Bertherat J. MANAGEMENT OF ENDOCRINE DISEASE: Adrenocortical carcinoma: differentiating the good from the poor prognosis tumors. Eur J Endocrinol 2018;178:R215-30. [Crossref] [PubMed]
- Tierney JF, Chivukula SV, Poirier J, et al. National Treatment Practice for Adrenocortical Carcinoma: Have They Changed and Have We Made Any Progress? J Clin Endocrinol Metab 2019;104:5948-56. [Crossref] [PubMed]
- Fassnacht M, Assie G, Baudin E, et al. Adrenocortical carcinomas and malignant phaeochromocytomas: ESMO-EURACAN Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol 2020;31:1476-90. [Crossref] [PubMed]
- Terzolo M, Fassnacht M, Perotti P, et al. Adjuvant mitotane versus surveillance in low-grade, localised adrenocortical carcinoma (ADIUVO): an international, multicentre, open-label, randomised, phase 3 trial and observational study. Lancet Diabetes Endocrinol 2023;11:720-30. [Crossref] [PubMed]
- Assié G, Jouinot A, Fassnacht M, et al. Value of Molecular Classification for Prognostic Assessment of Adrenocortical Carcinoma. JAMA Oncol 2019;5:1440-7. [Crossref] [PubMed]
- Yip L, Duh QY, Wachtel H, et al. American Association of Endocrine Surgeons Guidelines for Adrenalectomy: Executive Summary. JAMA Surg 2022;157:870-7. [Crossref] [PubMed]
- Sigala S, Bothou C, Penton D, et al. A Comprehensive Investigation of Steroidogenic Signaling in Classical and New Experimental Cell Models of Adrenocortical Carcinoma. Cells 2022;11:1439. [Crossref] [PubMed]
- Giordano TJ, Kuick R, Else T, et al. Molecular classification and prognostication of adrenocortical tumors by transcriptome profiling. Clin Cancer Res 2009;15:668-76. [Crossref] [PubMed]
- Guillaud-Bataille M, Ragazzon B, de Reyniès A, et al. IGF2 promotes growth of adrenocortical carcinoma cells, but its overexpression does not modify phenotypic and molecular features of adrenocortical carcinoma. PLoS One 2014;9:e103744. [Crossref] [PubMed]
- de Reyniès A, Assié G, Rickman DS, et al. Gene expression profiling reveals a new classification of adrenocortical tumors and identifies molecular predictors of malignancy and survival. J Clin Oncol 2009;27:1108-15. [Crossref] [PubMed]
- Mohan DR, Lerario AM, Else T, et al. Targeted Assessment of G0S2 Methylation Identifies a Rapidly Recurrent, Routinely Fatal Molecular Subtype of Adrenocortical Carcinoma. Clin Cancer Res 2019;25:3276-88. [Crossref] [PubMed]
- Vatrano S, Volante M, Duregon E, et al. Detailed genomic characterization identifies high heterogeneity and histotype-specific genomic profiles in adrenocortical carcinomas. Mod Pathol 2018;31:1257-69. [Crossref] [PubMed]
- Fassnacht M, Dekkers OM, Else T, et al. European Society of Endocrinology Clinical Practice Guidelines on the management of adrenocortical carcinoma in adults, in collaboration with the European Network for the Study of Adrenal Tumors. Eur J Endocrinol 2018;179:G1-G46. [Crossref] [PubMed]
- Berger MF, Mardis ER. The emerging clinical relevance of genomics in cancer medicine. Nat Rev Clin Oncol 2018;15:353-65. [Crossref] [PubMed]
- Tan YT, Lin JF, Li T, et al. LncRNA-mediated posttranslational modifications and reprogramming of energy metabolism in cancer. Cancer Commun (Lond) 2021;41:109-20. [Crossref] [PubMed]
- Winkle M, El-Daly SM, Fabbri M, et al. Noncoding RNA therapeutics - challenges and potential solutions. Nat Rev Drug Discov 2021;20:629-51. [Crossref] [PubMed]
- Detomas M, Pivonello C, Pellegrini B, et al. MicroRNAs and Long Non-Coding RNAs in Adrenocortical Carcinoma. Cells 2022;11:2234. [Crossref] [PubMed]
- Chifu I, Heinze B, Fuss CT, et al. Impact of the Chemokine Receptors CXCR4 and CXCR7 on Clinical Outcome in Adrenocortical Carcinoma. Front Endocrinol (Lausanne) 2020;11:597878. [Crossref] [PubMed]
- He J, Cao Y, Su T, et al. Downregulation of miR-375 in aldosterone-producing adenomas promotes tumour cell growth via MTDH. Clin Endocrinol (Oxf) 2015;83:581-9. [Crossref] [PubMed]
- Li W, Liu R, Wei D, et al. Circular RNA circ-CCAC1 Facilitates Adrenocortical Carcinoma Cell Proliferation, Migration, and Invasion through Regulating the miR-514a-5p/C22orf46 Axis. Biomed Res Int 2020;2020:3501451. [Crossref] [PubMed]
- Bhan A, Soleimani M, Mandal SS. Long Noncoding RNA and Cancer: A New Paradigm. Cancer Res 2017;77:3965-81. [Crossref] [PubMed]
- Yang M, Lu H, Liu J, et al. lncRNAfunc: a knowledgebase of lncRNA function in human cancer. Nucleic Acids Res 2022;50:D1295-306. [Crossref] [PubMed]
- Yuan K, Lan J, Xu L, et al. Long noncoding RNA TLNC1 promotes the growth and metastasis of liver cancer via inhibition of p53 signaling. Mol Cancer 2022;21:105. [Crossref] [PubMed]
- Siegel MB, He X, Hoadley KA, et al. Integrated RNA and DNA sequencing reveals early drivers of metastatic breast cancer. J Clin Invest 2018;128:1371-83. [Crossref] [PubMed]
- Zhang Z, Lu YX, Liu F, et al. lncRNA BREA2 promotes metastasis by disrupting the WWP2-mediated ubiquitination of Notch1. Proc Natl Acad Sci U S A 2023;120:e2206694120. [Crossref] [PubMed]
- Raju GSR, Pavitra E, Bandaru SS, et al. HOTAIR: a potential metastatic, drug-resistant and prognostic regulator of breast cancer. Mol Cancer 2023;22:65. [Crossref] [PubMed]
- Singh N, Ramnarine VR, Song JH, et al. The long noncoding RNA H19 regulates tumor plasticity in neuroendocrine prostate cancer. Nat Commun 2021;12:7349. [Crossref] [PubMed]
- Zhang R, Pan T, Xiang Y, et al. Curcumenol triggered ferroptosis in lung cancer cells via lncRNA H19/miR-19b-3p/FTH1 axis. Bioact Mater 2021;13:23-36. [Crossref] [PubMed]
- Zhang Y, Dong X, Guo X, et al. LncRNA-BC069792 suppresses tumor progression by targeting KCNQ4 in breast cancer. Mol Cancer 2023;22:41. [Crossref] [PubMed]
- Li S, Sun J, Ma J, et al. LncRNA LENGA acts as a tumor suppressor in gastric cancer through BRD7/TP53 signaling. Cell Mol Life Sci 2022;80:5. [Crossref] [PubMed]
- Palomares M, Delicado A, Mansilla E, et al. Characterization of a 8q21.11 microdeletion syndrome associated with intellectual disability and a recognizable phenotype. Am J Hum Genet 2011;89:295-301. [Crossref] [PubMed]
- Wang H, Niu L, Jiang S, et al. Comprehensive analysis of aberrantly expressed profiles of lncRNAs and miRNAs with associated ceRNA network in muscle-invasive bladder cancer. Oncotarget 2016;7:86174-85. [Crossref] [PubMed]
- Li SY, Wang H, Mai HF, et al. Down-regulated long non-coding RNA RNAZFHX4-AS1 suppresses invasion and migration of breast cancer cells via FAT4-dependent Hippo signaling pathway. Cancer Gene Ther 2019;26:374-87. [Crossref] [PubMed]
- Huang C, Cui H, Lang X, et al. An Integrative Analysis Revealing ZFHX4-AS1 as a Novel Prognostic Biomarker Correlated with Immune Infiltrates in Ovarian Cancer. J Immunol Res 2022;2022:9912732. [Crossref] [PubMed]
- Demeure MJ, Coan KE, Grant CS, et al. PTTG1 overexpression in adrenocortical cancer is associated with poor survival and represents a potential therapeutic target. Surgery 2013;154:1405-16; discussion 1416. [Crossref] [PubMed]
- Surakhy M, Wallace M, Bond E, et al. A common polymorphism in the retinoic acid pathway modifies adrenocortical carcinoma age-dependent incidence. Br J Cancer 2020;122:1231-41. [Crossref] [PubMed]
- Liberzon A, Birger C, Thorvaldsdóttir H, et al. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst 2015;1:417-25. [Crossref] [PubMed]
- Spearman's rank correlation coefficient. BMJ 2018;362:k4131. [PubMed]
- Wu T, Hu E, Xu S, et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation (Camb) 2021;2:100141. [Crossref] [PubMed]
- Mann HB, Whitney DR. On a Test of Whether one of Two Random Variables is Stochastically Larger than the Other. Ann Math Statist 1947;18:50-60. [Crossref]
- Ghosh C, Hu J, Kebebew E. Advances in translational research of the rare cancer type adrenocortical carcinoma. Nat Rev Cancer 2023;23:805-24. [Crossref] [PubMed]
- Fay AP, Elfiky A, Teló GH, et al. Adrenocortical carcinoma: the management of metastatic disease. Crit Rev Oncol Hematol 2014;92:123-32. [Crossref] [PubMed]
- Rodriguez-Galindo C, Krailo MD, Pinto EM, et al. Treatment of Pediatric Adrenocortical Carcinoma With Surgery, Retroperitoneal Lymph Node Dissection, and Chemotherapy: The Children's Oncology Group ARAR0332 Protocol. J Clin Oncol 2021;39:2463-73. [Crossref] [PubMed]
- Libé R, Borget I, Ronchi CL, et al. Prognostic factors in stage III-IV adrenocortical carcinomas (ACC): an European Network for the Study of Adrenal Tumor (ENSAT) study. Ann Oncol 2015;26:2119-25. [Crossref] [PubMed]
- Adnane S, Marino A, Leucci E. LncRNAs in human cancers: signal from noise. Trends Cell Biol 2022;32:565-73. [Crossref] [PubMed]
- Mattick JS, Rinn JL. Discovery and annotation of long noncoding RNAs. Nat Struct Mol Biol 2015;22:5-7. [Crossref] [PubMed]
- Moran VA, Perera RJ, Khalil AM. Emerging functional and mechanistic paradigms of mammalian long non-coding RNAs. Nucleic Acids Res 2012;40:6391-400. [Crossref] [PubMed]
- Zhou Y, Shao Y, Hu W, et al. A novel long noncoding RNA SP100-AS1 induces radioresistance of colorectal cancer via sponging miR-622 and stabilizing ATG3. Cell Death Differ 2023;30:111-24. [Crossref] [PubMed]
- Gudbjartsson DF, Walters GB, Thorleifsson G, et al. Many sequence variants affecting diversity of adult human height. Nat Genet 2008;40:609-15. [Crossref] [PubMed]
- Chen S, Diao J, Yue Z, et al. Identification and validation of ferroptosis-related genes and immune cell infiltration in thyroid associated ophthalmopathy. Front Genet 2023;14:1118391. [Crossref] [PubMed]
- Sabaie H, Moghaddam MM, Moghaddam MM, et al. Bioinformatics analysis of long non-coding RNA-associated competing endogenous RNA network in schizophrenia. Sci Rep 2021;11:24413. [Crossref] [PubMed]
- Guo J, Wu Y, Sun Y, et al. Bioinformatics-Based Screening of Key LncRNAs for Modulating the Transcriptome Associated with Glaucoma in Human Trabecular Meshwork Cells. Front Biosci (Landmark Ed) 2024;29:91. [Crossref] [PubMed]
- Boque-Sastre R, Guil S. A lncRNA Decoy Predicts Sensitivity to Cisplatin. Trends Mol Med 2020;26:352-4. [Crossref] [PubMed]
- Lin X, Zhuang S, Chen X, et al. lncRNA ITGB8-AS1 functions as a ceRNA to promote colorectal cancer growth and migration through integrin-mediated focal adhesion signaling. Mol Ther 2022;30:688-702. [Crossref] [PubMed]
- Xu J, Li Y, Lu J, et al. The mRNA related ceRNA-ceRNA landscape and significance across 20 major cancer types. Nucleic Acids Res 2015;43:8169-82. [Crossref] [PubMed]
- Chan JJ, Tay Y. Noncoding RNA:RNA Regulatory Networks in Cancer. Int J Mol Sci 2018;19:1310. [Crossref] [PubMed]
- Sherman MA, Yaari AU, Priebe O, et al. Genome-wide mapping of somatic mutation rates uncovers drivers of cancer. Nat Biotechnol 2022;40:1634-43. [Crossref] [PubMed]
- Assié G, Letouzé E, Fassnacht M, et al. Integrated genomic characterization of adrenocortical carcinoma. Nat Genet 2014;46:607-12. [Crossref] [PubMed]
- Yu F, Yu C, Li F, et al. Wnt/β-catenin signaling in cancers and targeted therapies. Signal Transduct Target Ther 2021;6:307. [Crossref] [PubMed]
- Gaujoux S, Grabar S, Fassnacht M, et al. β-catenin activation is associated with specific clinical and pathologic characteristics and a poor outcome in adrenocortical carcinoma. Clin Cancer Res 2011;17:328-36. [Crossref] [PubMed]
- Liu J, Xiao Q, Xiao J, et al. Wnt/β-catenin signalling: function, biological mechanisms, and therapeutic opportunities. Signal Transduct Target Ther 2022;7:3. [Crossref] [PubMed]
- Altieri B, Ronchi CL, Kroiss M, et al. Next-generation therapies for adrenocortical carcinoma. Best Pract Res Clin Endocrinol Metab 2020;34:101434. [Crossref] [PubMed]
- Crona J, Beuschlein F. Adrenocortical carcinoma - towards genomics guided clinical care. Nat Rev Endocrinol 2019;15:548-60. [Crossref] [PubMed]
- Barriga FM, Tsanov KM, Ho YJ, et al. MACHETE identifies interferon-encompassing chromosome 9p21.3 deletions as mediators of immune evasion and metastasis. Nat Cancer 2022;3:1367-85. [Crossref] [PubMed]
- Han G, Yang G, Hao D, et al. 9p21 loss confers a cold tumor immune microenvironment and primary resistance to immune checkpoint therapy. Nat Commun 2021;12:5606. [Crossref] [PubMed]
- Bui TO, Dao VT, Nguyen VT, et al. Genomics of Clear-cell Renal Cell Carcinoma: A Systematic Review and Meta-analysis. Eur Urol 2022;81:349-61. [Crossref] [PubMed]
- Koutros S, Kiemeney LA, Pal Choudhury P, et al. Genome-wide Association Study of Bladder Cancer Reveals New Biological and Translational Insights. Eur Urol 2023;84:127-37. [Crossref] [PubMed]
- Spiliopoulou P, Yang SYC, Bruce JP, et al. All is not lost: learning from 9p21 loss in cancer. Trends Immunol 2022;43:379-90. [Crossref] [PubMed]



