
Research on key pathogenesis and potential intervention targets of idiopathic renal calculi composed of calcium oxalate (CaOx) based on bioinformatics
Highlight box
Key findings
• This study utilized bioinformatics technology to analyze the gene expression profile chip data of idiopathic calcium oxalate (CaOx) kidney stones in depth, aiming to reveal their key pathogenesis and identify potential therapeutic targets.
What is known and what is new?
• CaOx kidney stones are the most common type in the urinary system with complex formation mechanisms.
• This study is the first to use bioinformatics methods to reveal the pathogenesis of idiopathic CaOx kidney stones at the molecular level and to identify the key gene interleukin 11 (IL-11) and its related signaling pathways. It further confirmed that the abnormal expression of IL-11 is closely associated with postoperative pyelonephritis, providing new insights for the prevention and treatment of postoperative infections.
What is the implication, and what should change now?
• Implications include the potential for IL-11 to be used in diagnostic and therapeutic strategies for idiopathic CaOx kidney stones.
• Further clinical trials are needed to validate IL-11 as a treatment target, potentially leading to new interventions and management approaches for patients with idiopathic CaOx kidney stones.
Introduction
It is estimated that more than 1–5% of the global population has a history of urolithiasis (1). In China, the incidence of kidney stones in adults is approximately 7.54% (2). The incidence rate is significantly higher in men than women (3). Calcium oxalate (CaOx) stones are the most common type of kidney stones, accounting for over 80% of kidney stones. The formation of kidney stones is a multifactorial and complex process involving the interaction of multiple aspects such as genetics, metabolism, environment, and lifestyle habits. The formation of CaOx stones in urine may be related to various factors such as pH value, urine volume, concentration of inhibitors and promoters in urine, and can usually be clearly displayed on X-rays (4). Apart from the rare cases in which single-gene genetic changes result in hereditary calcium renal calculi, and some CaOx stones secondary to gastrointestinal diseases, the majority of CaOx stones in clinical settings are idiopathic, lacking a clear genetic background and obvious etiological factors. The exact pathogenesis and molecular mechanisms of idiopathic CaOx stones remain largely unclear (5). Currently, the prevention and treatment of CaOx stones primarily rely on lifestyle modifications, medication, and surgical intervention (6). However, these treatments have limited efficacy and a high recurrence rate, significantly compromising the overall well-being of individuals. Therefore, exploring the key pathogenesis and potential intervention targets of CaOx kidney stones is of great clinical significance to improving treatment outcomes.
Research has shown that calcium phosphate crystals, also called Randall’s plaque (RP), deposited at the tips of the renal papillae in the renal medulla, serve as the initial site for kidney stone formation (7). Thus, the investigation of the key genes and signaling pathways involved in the formation of RP is essential to understand the pathogenesis of idiopathic renal calculi composed of CaOx. With the advent of high-throughput genomic research technologies, bioinformatics analysis software based on gene chip data has become an efficient tool for identifying and screening important genes related to the occurrence and progression of diseases (8). Our study used bioinformatics methods to analyze gene chip datasets from the public Gene Expression Omnibus (GEO) database to detect differentially expressed genes (DEGs) between kidney papilla tissues of idiopathic renal calculi composed of CaOx and normal kidney papilla tissues. This allowed us to screen the key genes and potential biochemical processes involved in the development of idiopathic renal calculi composed of CaOx, and thus identify novel targets and perspectives for its prevention and treatment. This study also retrospectively analyzed the clinical data of patients with idiopathic renal calculi composed of CaOx who underwent percutaneous nephrolithotomy (PCNL) to examine the expression of the key genes in their serum. By correlating the expression levels of these key genes with the incidence of idiopathic renal calculi composed of CaOx, we aimed to further clarify the clinical significance of these key genes. We present this article in accordance with the STREGA reporting checklist (available at https://tau.amegroups.com/article/view/10.21037/tau-24-302/rc).
Methods
Microarray data information
We retrieved the dataset GSE73680 using the keywords “idiopathic calcium oxalate stones” and “Randall’s plaques”, which compared the gene expression of renal papillary Randall plaques and normal tissues in 13 pairs of idiopathic CaOx stone forming individuals, annotations from the GEO database (https://www.ncbi.nlm.nih.gov/geo/) was used to label all identity documents (IDs) with corresponding gene symbols, and duplicate gene names were removed using the mean method.
Identification of DEGs
The limma package [3.52.2] (https://bioconductor.org/packages/release/bioc/html/limma.html) was used to screen the DEGs across all the datasets. Preprocess the raw data, including removing low-quality samples, correcting batch effects, standardizing gene expression levels, etc., to ensure data consistency and comparability. Provide necessary inputs for the linear model in the limma package based on the grouping information of the samples (i.e., RP renal papillary tissue formed by calcium stones and normal renal papillary tissue). Use the empirical Bayesian method in the limma package to statistically test the differences in gene expression levels. Use P<0.05 and log |fold change (FC)| >1 as screening criteria. DEGs with log FC >1 and log FC <−1 are considered upregulated and downregulated genes, respectively.
Identification of hub genes and establishment of protein-protein interaction (PPI) network
The Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database (https://cn.string-db.org/) was employed to evaluate the PPI network of the DEGs. Cytoscape v.3.9.1 (https://cytoscape.org/) was employed to visualize the positions and relationships of the key DEGs in the PPI network. The Molecular Complex Detection (MCODE) plugin in Cytoscape was used to identify central clusters to help identify potential functional modules and provide new insights into the activating signaling pathways.
Construction of transcription factor (TF)-DEG-microRNA (miRNA) network
The ENCODE (https://www.encodeproject.org/) and MiRTarBase v.8.0 databases (https://mirtarbase.cuhk.edu.cn/~miRTarBase/miRTarBase_2019/php/index.php) were used to acquire interaction data between the miRNAs-DEGs, and the TFs-DEGs. The DEGs displaying interactions with the TFs and miRNAs were chosen, after which the miRNAs and TFs regulating these DEGs were extracted. Cytoscape was used to visualize the integrated TF-DEG-miRNA network.
Gene set enrichment analysis (GSEA)
The Metascape online tool (https://metascape.org/gp/index.html) was employed to conduct the functional analysis of the DEGs. A GSEA was conducted to examine whether there were statistically significant differences in the biological expression of a predefined set of genes. By identifying common signaling pathways and regulatory network modules, this analysis provided insights into the biological differences between the different samples.
Data of study subjects
A retrospective analysis was conducted on the clinical information of 116 patients with idiopathic renal calculi composed of CaOx who underwent PCNL at General Hospital of Northern Theater Command from January 2020 to December 2022. These patients comprised the study group. To be eligible for inclusion in this study, the patients had to meet the following inclusion criteria: (I) meet the diagnostic criteria for urinary system stones as outlined in “Urology” by Jieping Wu (2004 edition) (9); (II) have confirmation that the main component of the postoperative stone specimens was CaOx based on an analysis with an infrared spectroscopy automatic analysis system; (III) be aged 18 years or older; (IV) have no history of gout, coronary heart disease, diabetes, autoimmune diseases, or hyperparathyroidism; and (V) have complete medical records. Patients were excluded from the study if they met any of the following exclusion criteria: (I) had co-existing diseases that could cause urinary stones, such as renal tubular acidosis, gout, nephrocalcinosis, primary cystinuria, hyperuricemia, or congenital urinary tract obstruction; (II) had severe acute urinary tract infection or gross hematuria; (III) had a severe trauma or surgical treatment within the past 6 months; (IV) had uncontrolled diabetes, liver or kidney dysfunction, hyperparathyroidism, cardiovascular diseases, cerebrovascular diseases, hematological diseases, autoimmune diseases, and/or malignant tumors; and/or (V) had severe mental or psychological disorders, or were non-cooperative with the follow-up. (VI) Merge chronic diseases that affect intestinal absorption and metabolism, such as Crohn’s disease, chronic diarrhea, short bowel syndrome, history of intestinal surgery. A control group of 110 healthy adults undergoing routine physical examinations during the same period was selected. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of General Hospital of Northern Theater Command (No. 2020-3) and informed consent was taken from all the participants.
Enzyme-linked immunosorbent assay (ELISA) assay
Fasting venous blood (5 mL) was collected from the elbow of all the participants using procoagulant/separation gel vacuum blood collection tubes. The samples were centrifuged at 3,500 r/min for 10 minutes at 4 ℃ to obtain the serum, which was then stored at −80 ℃. The expression levels of the key DEGs in the serum were measured using ELISA kits (Shanghai Zhenke Bio-Technology Co., Ltd., Shanghai, China).
Statistical analysis
GraphPad Prism 7.0 software (https://www.graphpad-prism.cn/) was used for the graphical plotting and statistical analysis. In the analysis of the DEGs, t-tests were used to determine the P values and adjusted P values, with the P values adjusted by the false discovery rate. The categorical data are presented as the number of cases (%). The groups were compared using the χ2 test. The Kolmogorov-Smirnov test was used to assess the normal distribution of the continuous data. Independent sample t-tests were applied to compare the normally distributed data, and the data are presented as the mean ± standard deviation (SD). The non-normally distributed data are expressed as the median and interquartile range [M (P25, P75)], and were compared using the Mann-Whitney U test. A binary logistic regression analysis was used to conduct the multivariate analysis. A P value less than 0.05 was considered statistically significant.
Results
Data normalization and identification of DEGs
We analyzed the gene expression of the GSE73680 dataset from the GEO database, focusing on the DEGs between RPs from CaOx stone-forming kidney papillae and normal kidney papillae tissues. Preprocessing included normalizing the data and performing cross-comparisons to remove technical and systemic variability. A principal component analysis (PCA) was performed to evaluate the biological variability between the samples. The PCA plot revealed unique gene expression profiles (Figure 1A), and the box plot illustrated the gene expression range for each sample (Figure 1B), which indicated the data suitability for further analysis. The raw data were robustly normalized to ensure the reliability of the downstream analyses. The limma package was used in R to identify the DEGs that met the following criteria: |log FC| >1 and P<0.05. In total, 276 upregulated and 538 downregulated genes were identified. The volcano plot of the DEGs (Figure 1C), and the heatmap (Figure 1D) revealed substantial differences in gene expression between the two groups.

Construction of PPI network and TF-DEG-miRNA network
Using the STRING database, we constructed a PPI network to illustrate the associations among the proteins encoded by the DEGs (Figure 2A). The majority of proteins encoded by the DEGs showed high interconnectivity with other proteins. The module analysis with MCODE identified the most notable module in the PPI network, which contained three genes (Figure 2B). The DEGs in this module were interleukin 11 (IL-11), interleukin 16 (IL-16), and interleukin 32 (IL-32). Among these, IL-11 was significantly differentially expressed between the two groups based on the GEO database (Figure 2C). To explore the functional roles of the DEGs, we explored the possible regulatory relationships between the TFs and DEGs, and also between the miRNAs and DEGs. Through a network analysis using the miRTarBase database, pairs of miRNA-DEGs were identified. The identification of potential regulatory links between the DEGs and TFs was based on the analysis of the TF binding site data. The genomic coordinates were provided by ENCODE (Figure 2D). Notably, IL-11 is potentially regulated by 25 TFs and interacts with six miRNAs.

Prediction of signaling pathways based on the GSEA
We conducted a functional GSEA of IL-11 using Metascape. The analysis revealed that IL-11 influences the occurrence and progression of idiopathic renal calculi composed of CaOx by affecting chemokine expression, and the signaling pathways of the toll-like receptors (TLRs) and nucleotide-binding oligomerization domain-like receptors [NOD-like receptors (NLRs)] (Figure 3).

Serum IL-11 gene expression levels in the two study groups
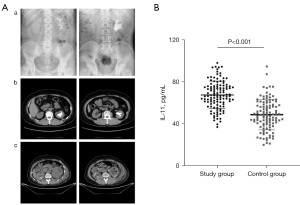
The study included imaging results of kidney stones from two patients as shown in Figure 4A. The ELISA results indicated that the IL-11 serum levels of the study group (67.44±13.15 pg/mL) were significantly increased compared to those of the control group (48.52±14.41 pg/mL) (P<0.001) (Figure 4B).
Analysis of factors influencing the occurrence of idiopathic renal calculi composed of CaOx
The univariate analysis showed that age, gender, body mass index (BMI), systolic blood pressure (SBP), total cholesterol (TC), low-density lipoprotein (LDL), and serum IL-11 levels were significantly associated with the occurrence of idiopathic renal calculi composed of CaOx (all P<0.05) (Table 1). The variables that were statistically significant in the univariate analysis were then included in the multivariate logistic regression model. The results indicated that gender, BMI, SBP, TC, and IL-11 were independent risk factors for the occurrence of idiopathic renal calculi composed of CaOx (all P<0.01) (Table 2).
Table 1
| Variables | Study group (n=116) | Control group (n=110) | t/χ2 | P value |
|---|---|---|---|---|
| Age (years) | 55.57±5.68 | 54.03±5.49 | 2.071 | 0.04 |
| Sex | 5.583 | 0.02 | ||
| Male | 75 | 54 | ||
| Female | 41 | 56 | ||
| BMI (kg/m2) | 23.02±1.04 | 22.34±0.87 | 5.317 | <0.001 |
| Smoking history | 2.223 | 0.14 | ||
| Yes | 64 | 45 | ||
| No | 52 | 55 | ||
| SBP (mmHg) | 101.19±15.34 | 94.95±14.14 | 3.175 | 0.002 |
| TC (mmol/L) | 2.15±0.89 | 1.86±0.73 | 2.670 | 0.008 |
| LDL (mmol/L) | 2.10±0.81 | 1.87±0.61 | 2.401 | 0.02 |
| IL-11 (pg/mL) | 67.44±13.15 | 48.52±14.41 | 10.32 | <0.001 |
Data are presented as mean ± standard deviation or n. BMI, body mass index; SBP, systolic blood pressure; TC, total cholesterol; LDL, low-density lipoprotein; IL-11, interleukin 11.
Table 2
| Variables | Exp (B) (95% CI) | P value |
|---|---|---|
| Age | 1.063 (0.996–1.134) | 0.07 |
| Sex (male) | 2.884 (1.321–6.299) | 0.008 |
| BMI | 2.147 (1.403–3.286) | <0.001 |
| SBP | 1.038 (1.011–1.066) | 0.006 |
| TC | 1.526 (0.982–2.372) | 0.06 |
| LDL | 1.118 (0.693–1.805) | 0.65 |
| IL-11 | 1.115 (1.080–1.151) | <0.001 |
Exp (B), exponential of the beta coefficient; CI, confidence interval; BMI, body mass index; SBP, systolic blood pressure; TC, total cholesterol; LDL, low-density lipoprotein; IL-11, interleukin 11.
Discussion
Recently, with the continuous advancement of molecular biology techniques, DNA microarrays, and next-generation sequencing technology, the investigation of DEGs of idiopathic renal calculi composed of CaOx at the transcriptome level and the analysis of key genes have become crucial methods for studying the risk factors and molecular mechanisms of this condition. Multiple studies have shown that the abnormal transcriptional activity of certain genes has a major effect on the formation and progression of idiopathic renal calculi composed of CaOx (10,11). Screening the differential gene expression profiles related to idiopathic CaOx kidney stones and identifying the key genes involved in their development is essential for the diagnosis, prevention, and treatment of this condition. In this study, by analyzing the GSE73680 dataset and using bioinformatics techniques, we identified 276 upregulated and 538 downregulated DEGs. Using the STRING database to construct a PPI network, three key genes were initially identified: IL-11, IL-16, and IL-32.
Previous research has shown that the interleukin family plays several vital roles in urinary stone formation. Specifically, the interleukin family: (I) promotes inflammatory responses: interleukins can trigger both local and systemic immune responses, contributing to tissue damage in the urinary tract and chronic low-grade systemic changes; (II) regulates calcium ion metabolism: certain interleukins may influence the absorption, excretion, or transport of calcium ions in the body, thereby increasing the risk of oxalate precipitates (such as oxalate) in the urine; (III) stimulates epithelial cell activation: when produced in the surrounding renal tissues, some interleukins can activate adjacent epithelial cells, enhancing their adhesive capacity required for nucleation and aggregation of crystal nuclei; and (IV) disrupts the fibrinolytic system balance: some interleukins can induce the deposition of various oxides, including the formation of calcium phosphate and other oxides, by interfering with the fibrinolytic system in an alkaline environment (12-14). The construction of the TF-DEG-miRNA network revealed that IL-11 alone is subject to TF regulation and potential miRNA interactions. Taguchi et al. (15) conducted a genome-wide analysis and found that the transcriptional activity of LCN2, IL-11, PTGS1, GPX3, and MMD is upregulated in RP tissues, which aid in the development of idiopathic renal calculi composed of CaOx, compared to non-RP tissues.
The GSEA suggested that IL-11 influences the occurrence and formation of idiopathic renal calculi composed of CaOx by affecting chemokine expression, and the signaling pathways of the TLR and NLR. These mechanisms are also supported by other studies in the context of kidney stones. For example, Gao et al. (16) used bioinformatics approaches to highlight the role of chemokine activity as a crucial pathway in kidney stone formation. Similarly, Olcucu et al. (17) demonstrated changes in subtypes of TLR in experimentally induced kidney stone disease, with real-time quantitative polymerase chain reaction showing a substantial decrease in expression of several TLRs, specifically TLR11 and TLR7. These findings are valuable for understanding the pathogenesis of idiopathic CaOx kidney stones. Current research on IL-11 primarily focuses on its effects on kidney function and fibrosis. It has been observed that IL-11 levels in the kidney increase with the progression of renal failure. Neutralizing IL-11 can reduce epithelial-mesenchymal transition, fibrosis, and inflammation, thereby improving kidney function (18). Further, the excessive expression of IL-11 can cause fibrosis in the kidneys and heart. Conversely, the deletion mutation of IL-11 receptor subunit alpha (IL-11Rα) can preserve organ functionality by preventing fibrosis (19). However, direct studies on IL-11 and kidney stones are limited (20), indicating a need for further research and validation in this area.
Through a retrospective analysis comparing serum IL-11 expression levels between patients with idiopathic renal calculi composed of CaOx and healthy subjects, this study preliminarily discovered that the expression of IL-11 was significantly upregulated in the serum of patients with idiopathic CaOx kidney stones. This suggests that IL-11 might play an important part in the pathological process of this condition. Further analysis revealed that sex, BMI, SBP, TC, and the expression level of IL-11 are independent risk factors for the occurrence of idiopathic renal calculi composed of CaOx. Study has shown that men are more prone to kidney stones than women (21), possibly because estrogen in women reduces calcium excretion, thereby lowering the risk of stone formation. Obesity increases the risk of kidney stones through various mechanisms, as excessive food intake can lead to metabolic imbalances of calcium, sodium, oxalate, and gastric acid, making obese individuals more susceptible to urinary stones (22). High blood pressure and high cholesterol can cause abnormal excretion of calcium and other minerals in the urine, promoting stone formation (23,24). IL-11 plays a significant role in kidney fibrosis, which may alter normal kidney function and promote stone formation. Considering these findings, IL-11 might serve as a diagnostic molecular marker and therapeutic target for idiopathic CaOx, offering a novel perspective for the diagnosis and treatment of this disease.
Conclusions
In summary, the development of idiopathic renal calcifications composed of CaOx is likely regulated by multiple factors. The identification of significant genes and related signal transduction pathways enhances our understanding of their molecular mechanisms. However, this study had certain limitations, including the analysis being restricted to single-center case data, and the lack of in vivo and in vitro mechanistic studies. Future research should involve multi-center, large-sample, prospective studies and validation through cellular or animal experiments to further explore and confirm the related findings.
Acknowledgments
Funding: This study was supported by
Footnote
Reporting Checklist: The authors have completed the STREGA reporting checklist. Available at https://tau.amegroups.com/article/view/10.21037/tau-24-302/rc
Data Sharing Statement: Available at https://tau.amegroups.com/article/view/10.21037/tau-24-302/dss
Peer Review File: Available at https://tau.amegroups.com/article/view/10.21037/tau-24-302/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tau.amegroups.com/article/view/10.21037/tau-24-302/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of General Hospital of Northern Theater Command (No. 2020-3) and informed consent was taken from all the participants.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Kachkoul R, Touimi GB, El Mouhri G, et al. Urolithiasis: History, epidemiology, aetiologic factors and management. Malays J Pathol 2023;45:333-52. [PubMed]
- Wang W, Fan J, Huang G, et al. Prevalence of kidney stones in mainland China: A systematic review. Sci Rep 2017;7:41630. [Crossref] [PubMed]
- Li ML, Song SC, Yang F, et al. Risk assessment and prevention of urolithiasis in urban areas of Baoding, China. Medicine (Baltimore) 2024;103:e35880. [Crossref] [PubMed]
- Shaltout AA, Dabi MM, Ahmed SI, et al. Spectroscopic Characterization of Urinary Stones Richening with Calcium Oxalate. Biol Trace Elem Res 2021;199:2858-68. [Crossref] [PubMed]
- Khan SR, Canales BK, Dominguez-Gutierrez PR. Randall's plaque and calcium oxalate stone formation: role for immunity and inflammation. Nat Rev Nephrol 2021;17:417-33. [Crossref] [PubMed]
- Hoa TQ, Nguyet TTM, Van Anh NT, et al. Evaluation of Factors Influenced on the Effectiveness of Percutaneous Nephrolithotomy. Med Arch 2024;78:33-8. [Crossref] [PubMed]
- Almeras C, Daudon M, Estrade V, et al. Classification of the renal papillary abnormalities by flexible ureteroscopy: evaluation of the 2016 version and update. World J Urol 2021;39:177-85. [Crossref] [PubMed]
- Sedlazeck FJ, Lee H, Darby CA, et al. Piercing the dark matter: bioinformatics of long-range sequencing and mapping. Nat Rev Genet 2018;19:329-46. [Crossref] [PubMed]
- Wu J. Wu Jieping Urology. Jinan: Shandong Science and Technology Press Co., Ltd.; 2004.
- Guerra A, Ticinesi A, Allegri F, et al. Idiopathic calcium nephrolithiasis with pure calcium oxalate composition: clinical correlates of the calcium oxalate dihydrate/monohydrate (COD/COM) stone ratio. Urolithiasis 2020;48:271-9. [Crossref] [PubMed]
- Arcidiacono T, Mingione A, Macrina L, et al. Idiopathic calcium nephrolithiasis: a review of pathogenic mechanisms in the light of genetic studies. Am J Nephrol 2014;40:499-506. [Crossref] [PubMed]
- Ma Q, Hou C, Cui H, et al. The serum levels and significance of MCSF, CF XIV, and IL-20 in patients with urinary calculi. Materials Express 2022;12:1588-91. [Crossref]
- Gan XG, Wang ZH, Xu HT. Mechanism of miRNA-141-3p in Calcium Oxalate-Induced Renal Tubular Epithelial Cell Injury via NLRP3-Mediated Pyroptosis. Kidney Blood Press Res 2022;47:300-8. [Crossref] [PubMed]
- Kim JY, Kim YS, Chang IH, et al. Interleukin-1β, calcium-sensing receptor, and urokinase gene polymorphisms in korean patients with urolithiasis. Korean J Urol 2011;52:340-4. [Crossref] [PubMed]
- Taguchi K, Hamamoto S, Okada A, et al. Genome-Wide Gene Expression Profiling of Randall's Plaques in Calcium Oxalate Stone Formers. J Am Soc Nephrol 2017;28:333-47. [Crossref] [PubMed]
- Gao Y, Liu D, Zhou H, et al. Identification of biomarkers and potential therapeutic targets of kidney stone disease using bioinformatics. World J Urol 2024;42:17. [Crossref] [PubMed]
- Olcucu MT, Teke K, Yalcin S, et al. Characterizing the Association Between Toll-like Receptor Subtypes and Nephrolithiasis With Renal Inflammation in an Animal Model. Urology 2018;111:238.e1-5. [Crossref] [PubMed]
- Widjaja AA, Shekeran SG, Adami E, et al. A Neutralizing IL-11 Antibody Improves Renal Function and Increases Lifespan in a Mouse Model of Alport Syndrome. J Am Soc Nephrol 2022;33:718-30. [Crossref] [PubMed]
- Corden B, Adami E, Sweeney M, et al. IL-11 in cardiac and renal fibrosis: Late to the party but a central player. Br J Pharmacol 2020;177:1695-708. [Crossref] [PubMed]
- Shen S, Wei J, Kang W, et al. Elucidating shared biomarkers and pathways in kidney stones and diabetes: insights into novel therapeutic targets and the role of resveratrol. J Transl Med 2023;21:491. [Crossref] [PubMed]
- Sorokin I, Mamoulakis C, Miyazawa K, et al. Epidemiology of stone disease across the world. World J Urol 2017;35:1301-20. [Crossref] [PubMed]
- Liu M, Wu J, Gao M, et al. Lifestyle factors, serum parameters, metabolic comorbidities, and the risk of kidney stones: a Mendelian randomization study. Front Endocrinol (Lausanne) 2023;14:1240171. [Crossref] [PubMed]
- Comellato G, Caletti C, Giani A, et al. Arterial stiffness and cardiovascular risk in patients with nephrolithiasis: a 10-year prospective study. J Hypertens 2024;42:1358-63. [Crossref] [PubMed]
- He Q, Tang Y, Li Y, et al. A pilot dynamic analysis of formative factors of nephrolithiasis related to metabolic syndrome: evidence in a rat model. Ren Fail 2022;44:1134-43. [Crossref] [PubMed]
(English Language Editor: L. Huleatt)


