
SPOP enhances FADD degradation and decreases the activeness of the NF-κB signaling pathway in prostate cancer: an in vitro study
Highlight box
Key findings
• Speckle-type POZ protein (SPOP) facilitates the degradation of FAS-associated protein with death domain (FADD), leading to a decrease in the activity of the nuclear transcription factor-κB (NF-κB) signaling pathway. There may be a SPOP-FADD-NF-κB regulatory axis in prostate cancer (PCa).
What is known, and what is new?
• SPOP, FADD, and NF-κB have been shown to be associated with the development of PCa. FADD has been shown to activate the NF-κB pathway to promote tumorigenesis. SPOP has been shown to enhance the breakdown of FADD and inhibit the function of the NF-κB signaling pathway in non-small cell lung cancer.
• SPOP enhances the degradation of FADD through ubiquitination, leading to a decrease in the activity of the NF-κB signaling pathway and inhibiting the progression of PCa. This indicates the presence of a potential regulatory axis involving SPOP, FADD, and NF-κB in PCa.
What is the implication, and what should change now?
• The above-mentioned proteins may become novel targets and tools for the diagnosis and treatment of PCa. The results of this study also need to be combined with PCa cases in the clinic for more in-depth research and discussion.
IntroductionOther Section
Prostate cancer (PCa) is one of the most common cancers, and its prevalence continues to increase year by year (1-3). It has a high incidence rate among men, and represents a threat to men’s health (1-3). Despite its slow progression, many PCa patients are in the middle to late stages at the time of their initial diagnosis, due to the absence of visible signs in the early stages, which leads to a poor prognosis and high mortality rates. PCa is likely to advance to castration-resistant PCa, which has a poor prognosis.
The emergence of genome sequencing on a molecular scale has sparked increasing interest in gene-targeted therapy for PCa, making it a prominent area of research in recent times. Speckle-type POZ protein (SPOP) is a high-frequency mutant gene in PCa (4-6). FAS-associated protein with death domain (FADD) is highly expressed in PCa (7). Nuclear transcription factor-κB (NF-κB) is one of the molecules involved in the development of PCa (8,9). FADD has been shown to activate the NF-κB pathway to promote tumorigenesis (10,11). SPOP has been shown to enhance the breakdown of FADD and inhibit the function of the NF-κB signaling pathway in non-small cell lung cancer (12). It is not yet known whether this regulatory mechanism exists in PCa. This study aimed to investigate and explore the role of SPOP in regulating FADD and the NF-κB signaling pathway in PCa and its mechanism. We present this article in accordance with the ARRIVE and MDAR reporting checklists (available at https://tau.amegroups.com/article/view/10.21037/tau-2024-701/rc).
MethodsOther Section
Research subjects
Cells
The human benign prostatic hyperplasia (BPH) cell line was selected from the RWPE-1 cell line, which was purchased from Sunncell Biotechnology Company (Wuhan, China). The human PCa cell lines were selected from the LNCaP and PC3 cell lines, which were purchased from Pricella Biotechnology Company (Wuhan, China).
Animals
A total of 18 Specific pathogen free grade Bagg albino/c (SPF-grade BALB/C) male nude mice, aged 4–6 weeks, and weighing 20±2 g, were obtained from the Shanghai Laboratory Animal Center. The Xinjiang Medical University Laboratory Animal Center maintained the mice under controlled conditions at a room temperature of 22±2 ℃, relative humidity of 40–70%, diurnal rhythms, and a closed and sterile environment. The mice had ad libitum access to sterile water and standard feed. A protocol was prepared before the study without registration. The animal experiments were approved by the Ethics Committee of Xinjiang Medical University Affiliated Tumor Hospital (approval No. K-2022010), in compliance with institutional guidelines for the care and use of animals.
Cell culture and expression of SPOP and FADD
The PC3 cell line was cultured with a mixture of Ham’s F-12K (GIBCO, California, USA), 10% fetal bovine serum (FBS) (Excell Bio, Shanghai, China), and 1% phosphate buffered saline (PBS) (GIBCO, California, USA). The LNCaP cell line was cultured using RPMI-1640 (GIBCO, California, USA), 10% FBS, and 1% PBS. The RWPE-1 cell line was cultured with keratinocyte serum-free medium (GIBCO, California, USA), 0.05 mg/mL bovine pituitary extract (BPE) (Absin Biotechnology Company, Shanghai, China), 5 ng/mL epidermal growth factor (EGF) (Abcam, Cambridge, UK), and 1% PBS. All cells were maintained in a constant temperature incubator with 5% carbon dioxide at 37 ℃. Western blot analysis was used to examine the levels of SPOP and FADD expression in the PC3, LNCaP, and RWPE-1 cells. Rabbit anti-SPOP antibody was purchased from Bioss Biotechnology Company (Beijing, China), and recombinant anti-FADD antibody was purchased from Abcam (Cambridge, UK). The PC3 cells were selected for subsequent experiments according to the test results.
Immunoprecipitation
Immunoprecipitation was performed using an immunoprecipitation kit (Abcam Co.). PC3 cells in the logarithmic growth phase were lysed and centrifuged, and the supernatant was divided into three groups. SPOP antibody was added to the experimental group (SPOP-IP), immunoglobulin G (IgG) antibody was added to the IgG control group (IgG-IP), and the input group (lysate) was stored for subsequent experiments. After the binding of the antibodies had been completed, the experimental and control samples were each treated with 40 µL of protein A/G microbead suspension. The samples were then centrifuged and washed to eliminate any non-specifically bound substances before the proteins that had attached to the beads were released. Subsequently, the three protein groups underwent western blot analysis to detect FADD expression.
SPOP gene silencing and overexpression
SPOP gene overexpression lentivirus and silencing lentivirus were purchased from Jikai Gene Company (Shanghai, China). The siSPOP primer sequence was 5'-GAGGTGAGTGTTGTGCAAGAT-3'; the siControl primer sequence was 5'-TTCTCCGAACGTGTCACGT-3'. The overexpression SPOP primer sequences were Forward Primer: 5'-GAGGATCCCCCCGGGTACCGGTCGCCACCATGGCGGAGCCGAGCGGC-3'; and Reverse Primer: 5'-TCACCATGGTGGCGACCGGGGCTGACACTCAACTGAGCA-3' (the italic markers are enzymatic sites). Following the determination of the best conditions for infecting lentivirus and the identification of puromycin-resistant cells, PC3 cell models with silenced and overexpressed SPOP were established using fluorescence quantitative polymerase chain reaction (PCR). The SPOP expression levels in these models were then analyzed: The proliferative activity of the cells was detected by Cell Counting Kit 8 (CCK-8) assay, apoptosis was measured by flow cytometry, and cell migration and invasion were evaluated by Transwell assay.
Expression of SPOP, FADD, P-P50, P-P65, and IκBα in the SPOP-silencing and SPOP-overexpression PC3 cells
Western blot was used to detect the expression of SPOP, FADD, Phosphorylated P50 (P-P50), Phosphorylated P65 (P-P65), and Inhibitor of NF-κB alpha (IκBα) in the SPOP-silencing and SPOP-overexpression PC3 cells, and IκBα antibody was purchased from Affinity Biosciences Company (Melbourne, Australia), while anti-NF-κB p105/p50 antibody and anti-NF-κB p65 antibody were purchased from Abcam (Cambridge, UK).
MG132 treatment
The 26S proteasome inhibitor MG132 inhibits the ubiquitinated degradation of proteins. The SPOP-overexpression PC3 cells were cultured by adding 10 mmol/L MG132 (TargetMol, Boston, USA) for 6 h. Western blot was then performed on the PC3 cells, SPOP-overexpression PC3 cells, and MG132-treated SPOP-overexpression PC3 cells to examine the expression of FADD.
Constructing a tumor animal model with SPOP overexpression, and detecting FADD, P-P50, P-P65, and IκBα expression
The 18 mice were randomly numbered and divided into the following three groups: the SPOP overexpression, overexpression airborne, and control groups (six mice per group). No significant differences in body weight were observed between the mice. The mice in SPOP overexpression group were injected unilaterally in the axilla subcutaneously with 100 µL of SPOP overexpression cell suspension (total number of cells: 2×106) and 100 µL of stromal gel, overexpression airborne group was injected with 100 µL of airborne cell suspension (total number of cells: 2×106) and 100 µL of stromal gel, control group was injected with 100 µL PC3 cell suspension (total number of cells: 2×106), and 100 µL stromal gel. Throughout the experiment, the long (L) axis and short (W) axis of the subcutaneous tumors in the nude mice were measured every 4 days using vernier calipers starting from the point of cell injection. To analyze any fluctuations in size over time, the tumor volume was calculated using the following formula: V = 1/2 × LW2. The animals were executed on day 28, and the tumors were removed and weighed. The tumor growth inhibition rate (IR) was calculated as follows: IR = (the average tumor weight of the control group/the average tumor weight of the experimental group) ×100%. Hematoxylin-eosin (HE) staining was then employed to detect the pathomorphological characteristics of each group’s tumors. Western blot was used to determine the expression levels of FADD, P-P65, P-P50 and IκBα proteins in the tumor tissues.
Statistical analysis
The measurement data that followed a normal distribution were presented as the mean and standard deviation (
ResultsOther Section
Expression of SPOP and FADD in different cells
SPOP had a relative expression level of 0.284±0.029 in PC3 cells, 0.418±0.028 in LNCaP cells, and 0.519±0.036 in RWPE-1 cells as observed by western blot. Notably, SPOP expression was lower in the PC3 cells than RWPE-1 cells (P<0.001), and lower in the LNCap cells than the RWPE-1 cells (P=0.008). The relative expression of FADD in the PC3 cells, LNCaP cells, and RWPE-1 cells was 0.668±0.104, 0.708±0.032, and 0.411±0.046, respectively. Thus, FADD expression was higher in the PC3 cells than the RWPE-1 cells (P=0.004), and higher in the LNCaP cells than the RWPE-1 cells (P=0.002) (Figure 1).

There is an interaction between SPOP and FADD in PC3 cells
Protein extracts from untreated PC3 cells were incubated with SPOP antibody and detected by co-immunoprecipitation. SPOP and FADD were immunoprecipitated, indicating that there was an interaction between them (Figure 2).

Construction and detection of PC3 cell models with SPOP gene silencing and overexpression (Figures 3-5)
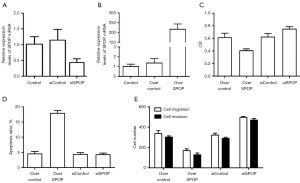
In the SPOP gene-silencing PC3 cell model, the cycle threshold (CT) values of SPOP expression in the control group, null-silencing group, and SPOP-silencing group, as detected by fluorescence quantitative PCR, were 1.017±0.235, 1.144±0.339, and 0.434±0.119, respectively. SPOP expression was lower in the silencing group than the control group (P=0.02), and it was also lower in the silencing group than the null-silencing group. (P=0.008) (Figure 3A). In the PC3 cell model with SPOP overexpression, fluorescence quantitative PCR revealed CT values of 1.017±0.235, 1.371±0.436, and 216.699±27.177 for the control, null, and overexpression groups, respectively. SPOP expression was higher in the overexpression group than the control group (P=0.02), and it was also higher in the overexpression group than the null-overexpression group (P=0.02) (Figure 3B).
The proliferative activity of the SPOP-silencing and SPOP-overexpression PC3 cells was detected by CCK-8, and the optical density (OD) values of the null-overexpression, overexpression, null-silencing, and silencing groups were 0.613±0.070, 0.405±0.029, 0.623±0.056, and 0.749±0.039, respectively. PC3 cell activity was higher in the SPOP null-overexpression group (P<0.001) than the SPOP-overexpression group, higher in the SPOP-silencing group than the null-silencing group (P=0.001), and even higher in the SPOP-silencing group than the SPOP-overexpression group (P<0.001) (Figure 3C).
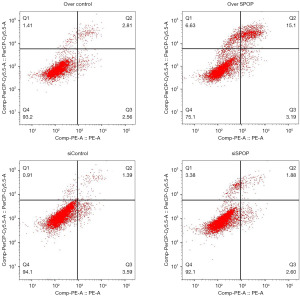
The apoptosis of the SPOP-silencing and SPOP-overexpression PC3 cells was detected by flow cytometry, and the apoptosis rates of the null-overexpression, overexpression, null-silencing and silencing groups were 4.520%±0.801%, 17.877%±0.913%, 4.340%±0.580%, and 4.253%±0.528%, respectively. The PC3 cell apoptosis rate was higher in the SPOP-overexpression group than the null-overexpression group (P<0.001), and even higher in the SPOP-overexpression group than the silencing group (P<0.001) (Figure 3D and Figure 4).
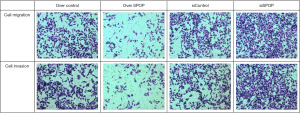
Transwell assay revealed cell migration and invasion. The cell migration numbers were 337.667±27.154, 169.333±17.214, 321.667±18.610, and 495.667±8.737 in the null-overexpression, SPOP-overexpression, null-silencing, and SPOP-silencing groups, respectively. The number of PC3 cells migrating was higher in the null-overexpression group than the SPOP-overexpression group (P<0.001), higher in the SPOP-silencing group than the null-silencing group (P<0.001), and even higher in the SPOP-silencing group than the SPOP-overexpression group (P<0.001) (Figure 3E and Figure 5).
The cell invasion numbers in the null-overexpression, SPOP-overexpression, null-silencing, and SPOP-silencing groups were 302.333±11.240, 128.333±17.616, 289.333±11.504, and 469.333±15.308, respectively. PC3 cell invasion was higher in the null-overexpression group than the SPOP-overexpression group (P<0.001), higher in the SPOP-silencing group than the null-silencing group (P<0.001), and higher in the SPOP-silencing group than the SPOP-overexpression group (P<0.001) (Figure 3E and Figure 5).
Expression of FADD, P-P50, P-P65, and IκBα in SPOP gene-silencing and SPOP gene-overexpression PC3 cells
The relative expression of FADD in the SPOP-overexpression, null-overexpression, SPOP-silencing, and null-silencing groups was 0.463±0.021, 0.585±0.027, 0.680±0.042, and 0.578±0.036, respectively. This indicates that the expression of FADD was lower in the SPOP-overexpression group than the null-overexpression group (P<0.001), and also lower in the SPOP-overexpression than the SPOP-silencing group (P<0.001). Conversely, FADD expression was higher in the SPOP-silencing group than the null-silencing group (P=0.001) (Figure 6A,6B).
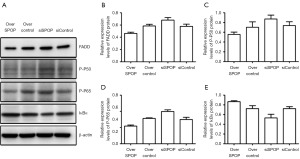
The relative expression of P-P50 in the SPOP-overexpression, null-overexpression, SPOP-silencing, and null-silencing groups was 0.555±0.049, 0.702±0.111, 0.871±0.080, and 0.736±0.080, respectively. This indicates that the level of P-P50 was lower in the SPOP-overexpression group than the null-overexpression group (P=0.03) and the SPOP-silencing group (P<0.001), while the expression of P-P50 was higher in the SPOP-silencing group than the null-silencing group (P=0.004) (Figure 6A,6C).
The relative expression of P-P65 in the SPOP-overexpression, null-overexpression, SPOP-silencing, and null-silencing groups was 0.544±0.032, 0.780±0.022, 0.998±0.051, and 0.749±0.068, respectively. This indicates that the expression of P-P65 was lower in the SPOP-overexpression group than the null-overexpression group (P<0.001) and the SPOP-silencing group (P<0.001), while the expression of P-P65 was higher in the SPOP-silencing group than the null-silencing group (P<0.001) (Figure 6A,6D).
The relative expression of IκBα in the SPOP-overexpression, null-overexpression, SPOP-silencing, and null-silencing groups was 0.864±0.021, 0.723±0.062, 0.531±0.075, and 0.722±0.040, respectively. This indicates that the expression of IκBα was higher in the SPOP-overexpression group than the null-overexpression group (P=0.003), and was also higher than in the silencing group (P<0.001). The expression of IκBα was significantly lower in the SPOP-silencing group than the null-silencing group (P<0.001) (Figure 6A,6E).
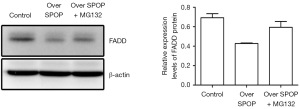
Effect of MG132 inhibitor on FADD expression in PC3 with cells overexpressing the SPOP gene
The relative expression of FADD in the PC3 cells, PC3 cells overexpressing the SPOP gene, and MG132-treated PC3 cells overexpressing the SPOP gene was 0.692±0.042, 0.428±0.005, and 0.596±0.060, respectively, as detected by western blot. The expression of FADD was lower in the SPOP-overexpression PC3 cells than the PC3 cells (P<0.001), and was also lower in the SPOP-overexpression PC3 cells than the MG132-treated SPOP-overexpression PC3 cells (P=0.003) (Figure 7).
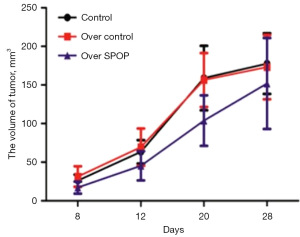
Construction of PCa tumor animal models with SPOP gene overexpression
Tumor volume was measured and calculated in the SPOP-overexpression, null-overexpression, and control groups (Figure 8). The animals were executed on the 28th day of modeling (Figure 9). The tumors were weighed, and the three groups had weights of 0.241±0.031, 0.295±0.033, and 0.305±0.037 g, respectively. The weight of the SPOP-overexpression group was lower than that of the null group (P=0.02) and the control group (P=0.005); the tumor growth inhibition rates for each group were 20.798%, 3.131%, and 0%, respectively.

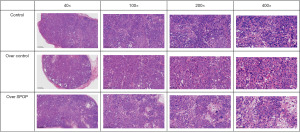
The results of the HE staining for the tumors showed that the tumor cells in the control group were densely packed when viewed under the microscope, with large nuclei that were deeply stained, prominent nucleoli, visible nuclear atypia, and a lack of intercellular bridges and cellular keratinization. Conversely, there was no notable variance in the level of tumor cell necrosis between the null-overexpression group and the control group. In comparison to the control and null-overexpression groups, some of the tumor cells in the SPOP-overexpression group were broken down and necrotic; large necrotic zones were visible in the center and around the tumor tissues, with a broadening of tumor necrosis (Figure 10).
Expression of FADD, P-P50, P-P65, and IκBα in PCa tumor animal models with SPOP gene-overexpression
Tumor protein expression was detected by western blot in the SPOP gene overexpression animal models, and the relative expression of FADD in the SPOP-overexpression, null-overexpression, and control groups were 0.390±0.021, 0.549±0.047, and 0.586±0.014, respectively. These results indicated that the expression of FADD was lower in the tumors of the SPOP-overexpression group than those of the null-overexpression group (P=0.001) and control group (P<0.001) (Figure 11A,11B).
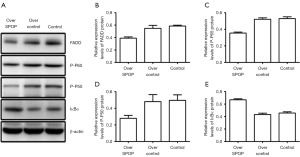
In the SPOP-overexpression group, the relative expression of P-P65 was 0.355±0.011, while in the null-overexpression group and control group, it was 0.521±0.018 and 0.529±0.022, respectively. These results indicated that the expression of P-P65 was significantly lower in the tumors of the SPOP-overexpression group than those of the null-overexpression group (P<0.001) and the control group (P<0.001) (Figure 11A,11C).
The relative expression of P-P50 in the SPOP-overexpression, null-overexpression, and control groups was 0.280±0.035, 0.481±0.084, and 0.495±0.067, respectively. These results indicated that the expression of P-P50 was lower in the tumors of the SPOP-overexpression group than those of both the null-overexpression group (P=0.009) and the control group (P=0.007) (Figure 11A,11D).
The levels of IκBα expression were found to be 0.669±0.015 in the SPOP-overexpression group, 0.434±0.021 in the null-overexpression group, and 0.456±0.020 in the control group. These results indicated that the IκBα expression was significantly higher in the tumors of the SPOP-overexpression group than those of both the null-overexpression group (P<0.001) and the control group (P<0.001) (Figure 11A,11E).
DiscussionOther Section
The ubiquitin proteasome pathway is a critical post-translational alteration process involved in the control of multiple cellular activities, such as cell proliferation, differentiation, transcription, and apoptosis (13). This same pathway plays an essential role in the development of tumors; if certain oncogene proteins are not removed from cells quickly enough by the proteasome, they can lead to malignancy. SPOP is the splice protein of the Cullin3-associated E3 ubiquitin ligase, and it exerts an antitumor effect by mediating the ubiquitination and degradation of a variety of pro-tumorigenic factors (14,15). The main SPOP acting substrates include Androgen receptor (AR), Steroid receptor coactivator 3 (SRC-3), and ETS-related gene (ERG) (16-18). Shi et al. found that cytoplasmically localized SPOP binds and mediates a non-degradative ubiquitination modification of the K420 site of the p62UBA structural domain. This modification reduced the ubiquitin-binding capacity of p62 and its spot formation and liquid phase separation, thereby inhibiting p62-dependent autophagy. Moreover, they show that SPOP inhibits the compartmentalization of Keap1 by p62, which ultimately reduces the transcriptional activation of antioxidant genes by Nrf2. In contrast, PCa-derived SPOP mutants lose the ability to ubiquitinate p62 and promote autophagy and redox reactions in a dominant-negative manner (19). FADD is a vital protein that connects the tumor necrosis factor receptor family with caspase-8/10 kininogen to create a death-inducing signaling complex in apoptosis. The signals triggering apoptosis are transmitted by the main death receptor into the cell, influencing various physiological and pathological processes, such as cell proliferation, tumor formation, cell cycle advancement, autophagy, inflammation, and natural immunity (20-22). NF-κB is a pleiotropic transcription factor belonging to the Rel/NF-κB family, which is involved in the activation of many genes, including cytokines and metalloproteinases (23,24).The NF-κB signaling pathway is crucial in numerous biological and disease-related processes, including cell proliferation, apoptosis, inflammatory response, immune response, tumorigenesis, and development (25,26). The linear ubiquitin chain assembly complex (LUBAC), which uses E3 ubiquitin ligase, is responsible for producing a linear ubiquitin chain and triggering the activation of NF-κB (27). Goto et al. found that FADD is also a protein substrate for ubiquitination by LUBAC (28). A large-scale protein interaction study reported a possible interaction between SPOP and FADD (29). Previous research had not examined whether a SPOP-FADD-NF-κB regulatory axis exists in PCa.
In this study, we first verified that the PCa cells had reduced SPOP expression and elevated FADD expression compared to the BPH cells. Further, we constructed SPOP gene-silencing and SPOP gene-overexpression PC3 cell models, and found that the proliferative activity, migration, and invasion of the SPOP-overexpression PC3 cells were lower than those of the SPOP-silencing PC3 cells, and the apoptosis rate of the SPOP-overexpression PC3 cells was higher than that of the SPOP-silencing PC3 cells. The levels of FADD and NF-κB were more elevated in the PC3 cells in the SPOP gene-silencing group than the PC3 cells in the null-silent group. Conversely, the levels of FADD and NF-κB were more reduced in the PC3 cells in the SPOP-overexpression group than in the PC3 cells in the null-overexpressed group. These results suggest that SPOP suppresses the proliferation of PC3 cells, while FADD and NF-κB stimulate the growth of PC3 cells.
A negative correlation was found between the expression of SPOP, FADD, and NF-κB. Subsequently, this study confirmed an interaction between SPOP and FADD in the PC3 cells by immunoprecipitation. Next, the PC3 cells overexpressing SPOP were treated with MG132, and the expression of FADD in the cells rebounded compared with those untreated with MG132, which indicated that the inhibition of the expression of FADD by SPOP was mediated by its ubiquitylation and degraded by 26S protease. A PCa animal model with SPOP gene overexpression was established, and the results showed that the expression of SPOP inhibited the growth of PCa tumors, and there was a negative correlation between SPOP, FADD, and NF-κB.
In this study, we explored and analyzed the mechanism by which SPOP regulates the NF-κB signaling pathway through its action on FADD in PCa for the first time. We found that SPOP has the ability to enhance the degradation of FADD through ubiquitination, leading to a decrease in the activity of the NF-κB signaling pathway and inhibiting the progression of PCa. This indicates the presence of a potential regulatory axis involving SPOP, FADD, and NF-κB in PCa. Differences in SPOP expression have been found to have guiding significance in the treatment and prognosis of PCa (30-35), but the relevant mechanisms have yet to be closely investigated. Our findings offer fresh perspectives and guidance for investigating PCa pathogenesis, and lay a new foundation for the diagnosis, treatment, and prevention of PCa. The related proteins in the study may become novel targets and tools for the diagnosis and treatment of PCa. It may be possible to block the expression of a certain gene in the SPOP-FADD-NF-κB signaling pathway through some kind of treatment to inhibit the progression of PCa. The conclusions of this study also need to be combined with PCa cases in the clinic for more in-depth research and discussion.
ConclusionsOther Section
SPOP facilitates the degradation of FADD, leading to a decrease in the activity of the NF-κB signaling pathway. There may be a SPOP-FADD-NF-κB regulatory axis in PCa.
AcknowledgmentsOther Section
Funding: This work was supported by
FootnoteOther Section
Reporting Checklist: The authors have completed the ARRIVE and MDAR reporting checklists. Available at https://tau.amegroups.com/article/view/10.21037/tau-2024-701/rc
Data Sharing Statement: Available at https://tau.amegroups.com/article/view/10.21037/tau-2024-701/dss
Peer Review File: Available at https://tau.amegroups.com/article/view/10.21037/tau-2024-701/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tau.amegroups.com/article/view/10.21037/tau-2024-701/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The animal experiments were approved by the Ethics Committee of Xinjiang Medical University Affiliated Tumor Hospital (approval No. K-2022010), in compliance with institutional guidelines for the care and use of animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
ReferencesOther Section
- Preisser F, Bandini M, Nazzani S, et al. Development and Validation of a Lookup Table for the Prediction of Metastatic Prostate Cancer According to Prostatic-specific Antigen Value, Clinical Tumor Stage, and Gleason Grade Groups. Eur Urol Oncol 2020;3:631-9. [Crossref] [PubMed]
- Elazab IM, El-Feky OA, Khedr EG, et al. Prostate cancer and the cell cycle: Focusing on the role of microRNAs. Gene 2024;928:148785. [Crossref] [PubMed]
- Jin W, Fei X, Wang X, et al. Circulating miRNAs as Biomarkers for Prostate Cancer Diagnosis in Subjects with Benign Prostatic Hyperplasia. J Immunol Res 2020;2020:5873056. [Crossref] [PubMed]
- Orme JJ, Taza F, De Sarkar N, et al. Co-occurring BRCA2/SPOP Mutations Predict Exceptional Poly (ADP-ribose) Polymerase Inhibitor Sensitivity in Metastatic Castration-Resistant Prostate Cancer. Eur Urol Oncol 2024;7:877-87. [Crossref] [PubMed]
- Segalés L, Juanpere N, Gallarín N, et al. Immunohistochemical markers as predictors of prognosis in multifocal prostate cancer. Virchows Arch 2024;485:281-90. [Crossref] [PubMed]
- Akhoundova D, Francica P, Rottenberg S, et al. DNA Damage Response and Mismatch Repair Gene Defects in Advanced and Metastatic Prostate Cancer. Adv Anat Pathol 2024;31:61-9. [Crossref] [PubMed]
- Wang C, Jiang X, Zhao Q, et al. The diagnostic or prognostic values of FADD in cancers based on pan‑cancer analysis. Biomed Rep 2023;19:77. [Crossref] [PubMed]
- Clark A, Villarreal MR, Huang SB, et al. Targeting S6K/NFκB/SQSTM1/Polθ signaling to suppress radiation resistance in prostate cancer. Cancer Lett 2024;597:217063. [Crossref] [PubMed]
- Yu J, Zhang M, Li T, et al. Monoacylglycerol lipase blockades the senescence-associated secretory phenotype by interfering with NF-κB activation and promotes docetaxel efficacy in prostate cancer. Oncogene 2024;43:2835-49. [Crossref] [PubMed]
- Ranjan K, Pathak C. Cellular Dynamics of Fas-Associated Death Domain in the Regulation of Cancer and Inflammation. Int J Mol Sci 2024;25:3228. [Crossref] [PubMed]
- Zeng X, Chen Z, Zhu Y, et al. O-GlcNAcylation regulation of RIPK1-dependent apoptosis dictates sensitivity to sunitinib in renal cell carcinoma. Drug Resist Updat 2024;77:101150. [Crossref] [PubMed]
- Luo J, Chen B, Gao CX, et al. SPOP promotes FADD degradation and inhibits NF-κB activity in non-small cell lung cancer. Biochem Biophys Res Commun 2018;504:289-94. [Crossref] [PubMed]
- Zhang P, Gao K, Jin X, et al. Endometrial cancer-associated mutants of SPOP are defective in regulating estrogen receptor-α protein turnover. Cell Death Dis 2015;6:e1687. [Crossref] [PubMed]
- Gebrael G, Zengin Z, Swami U. Differential Impact of SPOP Mutation in Prostate and Endometrial Cancers. JCO Precis Oncol 2023;7:e2300397. [Crossref] [PubMed]
- Yang Y, Han YC, Cao Q, et al. SPOP negatively regulates mTORC1 activity by ubiquitinating Sec13. Cell Signal 2024;116:111060. [Crossref] [PubMed]
- Wang Z, Song Y, Ye M, et al. The diverse roles of SPOP in prostate cancer and kidney cancer. Nat Rev Urol 2020;17:339-50. [Crossref] [PubMed]
- Zhang H, Jin X, Huang H. Deregulation of SPOP in Cancer. Cancer Res 2023;83:489-99. [Crossref] [PubMed]
- Hatano K, Nonomura N. Genomic Profiling of Prostate Cancer: An Updated Review. World J Mens Health 2022;40:368-79. [Crossref] [PubMed]
- Shi Q, Jin X, Zhang P, et al. SPOP mutations promote p62/SQSTM1-dependent autophagy and Nrf2 activation in prostate cancer. Cell Death Differ 2022;29:1228-39. [Crossref] [PubMed]
- Davidovich P, Higgins CA, Najda Z, et al. cFLIP(L) acts as a suppressor of TRAIL- and Fas-initiated inflammation by inhibiting assembly of caspase-8/FADD/RIPK1 NF-κB-activating complexes. Cell Rep 2023;42:113476. [Crossref] [PubMed]
- Ahanin EF, Sager RA, Backe SJ, et al. Catalytic inhibitor of Protein Phosphatase 5 activates the extrinsic apoptotic pathway by disrupting complex II in kidney cancer. Cell Chem Biol 2023;30:1223-1234.e12. [Crossref] [PubMed]
- Damhofer H, Tatar T, Southgate B, et al. TAK1 inhibition leads to RIPK1-dependent apoptosis in immune-activated cancers. Cell Death Dis 2024;15:273. [Crossref] [PubMed]
- Chen L, De Menna M, Groenewoud A, et al. A NF-κB-Activin A signaling axis enhances prostate cancer metastasis. Oncogene 2020;39:1634-51. [Crossref] [PubMed]
- Torrealba N, Vera R, Fraile B, et al. TGF-β/PI3K/AKT/mTOR/NF-kB pathway. Clinicopathological features in prostate cancer. Aging Male 2020;23:801-11. [Crossref] [PubMed]
- Minciacchi VR, Karantanou C, Bravo J, et al. Differential inflammatory conditioning of the bone marrow by acute myeloid leukemia and its impact on progression. Blood Adv 2024;8:4983-96. [Crossref] [PubMed]
- Strzyga-Łach P, Kurpios-Piec D, Chrzanowska A, et al. 1,3-Disubstituted thiourea derivatives: Promising candidates for medicinal applications with enhanced cytotoxic effects on cancer cells. Eur J Pharmacol 2024;982:176885. [Crossref] [PubMed]
- Chen YG, Rieser E, Bhamra A, et al. LUBAC enables tumor-promoting LTβ receptor signaling by activating canonical NF-κB. Cell Death Differ 2024;31:1267-84. [Crossref] [PubMed]
- Goto E, Tokunaga F. Decreased linear ubiquitination of NEMO and FADD on apoptosis with caspase-mediated cleavage of HOIP. Biochem Biophys Res Commun 2017;485:152-9. [Crossref] [PubMed]
- Ewing RM, Chu P, Elisma F, et al. Large-scale mapping of human protein-protein interactions by mass spectrometry. Mol Syst Biol 2007;3:89. [Crossref] [PubMed]
- Swami U, Graf RP, Nussenzveig RH, et al. SPOP Mutations as a Predictive Biomarker for Androgen Receptor Axis-Targeted Therapy in De Novo Metastatic Castration-Sensitive Prostate Cancer. Clin Cancer Res 2022;28:4917-25. [Crossref] [PubMed]
- Pedrani M, Salfi G, Merler S, et al. Prognostic and Predictive Role of SPOP Mutations in Prostate Cancer: A Systematic Review and Meta-analysis. Eur Urol Oncol 2024;7:1199-215. [Crossref] [PubMed]
- Wang D, Pan H, Cheng S, et al. Construction and Validation of a Prognostic Model Based on Mitochondrial Genes in Prostate Cancer. Horm Metab Res 2024;56:807-17. [Crossref] [PubMed]
- Bidot S, Yin J, Zhou P, et al. Genetic Profiling of African American Patients With Prostatic Adenocarcinoma Metastatic to the Lymph Nodes: A Pilot Study. Arch Pathol Lab Med 2024;148:310-7. [Crossref] [PubMed]
- Kumar AA. Prostate cancer genotyping for risk stratification and precision treatment. Curr Urol 2024;18:87-97. [Crossref] [PubMed]
- Sutera P, Song Y, Shetty AC, et al. Genomic Determinants Associated with Modes of Progression and Patterns of Failure in Metachronous Oligometastatic Castration-sensitive Prostate Cancer. Eur Urol Oncol 2024; Epub ahead of print. [Crossref] [PubMed]
(English Language Editor: L. Huleatt)


