
In silico research of coagulation- and fibrinolysis-related genes for predicting prognosis of clear cell renal cell carcinoma
Highlight box
Key findings
• This study established a prognostic model consisting of TIMP1, RUNX1, BMP6, and PROS1, coagulation- and fibrinolysis-related genes that play a significant role in clear cell renal cell carcinoma (ccRCC) progression. This model exhibits robust predictive capacity for ccRCC prognosis and underscores potential therapeutic targets for future treatment strategies.
What is known and what is new?
• TIMP1, RUNX1, BMP6, and PROS1 are associated with coagulation and fibrinolysis, and alterations in their expression may serve as indicators of disease progression and prognosis.
• A prognostic model was built based on coagulation and fibrinolysis-related genes in ccRCC through bioinformatics analysis and experimental validation.
What is the implication, and what should change now?
• The pathogenesis of renal clear cell carcinoma was explored, offering potential directions for the treatment of ccRCC.
• We would continue to take care of the research progress of the coagulation and fibrinolytic system and ccRCC, in expecting to have more treatment options based on this study.
Introduction
Background
Renal cell carcinoma (RCC) is a prevalent malignancy within the genitourinary system, comprising approximately 2.7% of new cancer cases in males and 1.6% in females globally in 2022, leading to 434,419 new diagnoses and 155,702 deaths (1). The primary subtypes of RCC include clear cell RCC (ccRCC), chromophobe RCC, and papillary RCC, with ccRCC being the most common (2,3). Prognosis in ccRCC varies significantly by stage; early-stage localized tumors have a 5-year survival rate exceeding 90%, while metastatic disease is associated with a survival rate as low as 12% at 5 years (4). Notably, from the late 1980s to 2006, the 5-year survival rate for stage IV patients remained below 10%, with a median overall survival of only 10–15 months (5). Currently, the standard treatment for advanced ccRCC involves a combination of targeted therapies and immunotherapy (6-8). Several drugs have demonstrated survival benefits in advanced patients through clinical trials (9,10), but substantial improvements remain necessary. The identification of prognostic genes in kidney cancer has gained considerable attention (11), and various predictive models, such as the Royal Marsden Hospital (RMH) Score, have been developed to assess prognostic outcomes (12). However, due to the poor prognosis of advanced ccRCC and the limited understanding of tumor-related prognostic genes, novel biomarkers are urgently needed to guide diagnosis and treatment strategies.
Rationale and knowledge gap
Cancerous tumors induce a hypercoagulable state by promoting inflammatory mediators and cytokines that activate the coagulation system. These mediators stimulate the synthesis and release of coagulation factors, such as tissue factor (TF) and thrombin. Tumors are often associated with vascular abnormalities, including neovascularization and increased vascular permeability, which may lead to thrombosis and further activation of the coagulation cascade. Subsequently, anticoagulation mechanisms are triggered in an attempt to restore homeostasis (13). Additionally, components of the fibrinolytic system, including tissue-type plasminogen activator (tPA), urokinase-type plasminogen activator (uPA), uPA receptor (uPAR), and plasminogen activator inhibitor 1 (PAI-1), are integral in modulating tumor development, proliferation, invasion, and metastasis (14). Overexpression of uPA in breast (15), colorectal (16), lung (17), and basal cell carcinoma (18) tissues has been linked to poor prognosis in patients with cancers (19-21). Extensive evidence highlights a direct and significant relationship between coagulation- and fibrinolysis-related genes (CFRGs) and cancer progression, suggesting potential therapeutic pathways across different cancer types (13,22-24). However, the precise action mechanisms of CFRGs in ccRCC remain poorly understood and warrant further investigation.
Objective
This study identifies genes associated with the coagulation and fibrinolysis systems in ccRCC, exploring their potential roles in tumor pathogenesis and progression. A risk assessment model was constructed to predict patient outcomes in ccRCC. Furthermore, immune microenvironment analysis revealed distinct immune cell infiltration patterns and regulatory mechanisms, offering new targets for immunotherapy. The construction of a long non-coding RNA-microRNA-messenger RNA (lncRNA-miRNA-mRNA) network provides fresh insights into the pathogenesis of ccRCC, while drug predictive analyses highlight potential treatment options, offering valuable guidance for clinical practice. We present this article in accordance with the TRIPOD reporting checklist (available at https://tau.amegroups.com/article/view/10.21037/tau-24-483/rc).
Methods
Collection of data
The Cancer Genome Atlas-Kidney Renal Clear Cell Carcinoma (TCGA-KIRC) dataset, comprising 522 ccRCC samples with survival data and 71 control samples, was obtained from the University of California Santa Cruz (UCSC) Xena (http://xena.ucsc.edu/). Additionally, 101 ccRCC samples from the E-MTAB-1980 cohort were retrieved from the ArrayExpress database (https://www.ebi.ac.uk/arrayexpress/). The GSE53757 dataset, including 72 ccRCC samples and 72 control individuals, was sourced from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/gds), with sample types consisting of flash-frozen patient disease and control samples, and data generated using the Affymetrix Human Genome U133 Plus 2.0 Array platform. Furthermore, 222 CFRGs were extracted from the GeneCards database (https://www.genecards.org/) using the keywords “coagulation” and “fibrinolysis” with a relevance score >3.
Identification of differentially expressed CFRGs (DE-CFRGs)
Differentially expressed genes (DEGs) between ccRCC and control cohorts in the TCGA-KIRC dataset were identified using the DESeq2 package (version 1.36.0) (25) with thresholds of |log2fold change (FC)| >1 and adjusted P<0.05. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses were performed using the clusterProfiler package (version 4.7.1.001) (26) to explore the biological processes and pathways associated with the DEGs (adj. P<0.05). Additionally, ccRCC and control samples from the TCGA-KIRC dataset were designated as traits for weighted gene co-expression network analysis (WGCNA). Outlier samples were excluded based on clustering analysis to ensure the accuracy of the results. A soft threshold was determined to maximize the scale-free distribution of gene interactions. Subsequently, a systematic clustering tree was constructed based on the dissimilarity coefficient, calculated from the adjacency and similarity of genes. Modules were then identified using dynamic tree cutting criteria. Pearson correlations between modules and traits were calculated, and the module most strongly correlated with ccRCC was selected as the key module. Finally, DEGs, CFRGs, and genes within the key module were overlapped to identify DE-CFRGs, followed by GO and KEGG enrichment analyses (adj. P<0.05).
Establishment of a prognostic model
Univariate Cox regression analysis was performed on DE-CFRGs using the expression matrix and survival data from the TCGA-KIRC dataset to identify genes associated with the survival of patients with ccRCC (HR ≠1 and P<0.05). Subsequently, least absolute shrinkage and selection operator (LASSO) analysis was applied to select genes corresponding to the minimum lambda (λmin), followed by multivariate Cox regression analysis to pinpoint prognostic genes (HR ≠1 and P<0.05). A risk model was then constructed, with the risk score for each ccRCC sample in TCGA-KIRC calculated as follows: risk score = α1 × X1 + α2 × X2 + … + αn × Xn, where α represents the coefficient and X denotes the relative gene expression. Based on the median risk score, ccRCC samples were divided into high- and low-risk groups. Kaplan-Meier (K-M) survival analysis was conducted to compare survival differences between these groups. The predictive ability of the risk model was evaluated using receiver operating characteristic (ROC) curves. Further validation of the model was carried out using the same methods in the E-MTAB-1980 cohort. Additionally, the expression levels of prognostic genes were compared between ccRCC and control samples using the Wilcoxon test in both the TCGA-KIRC and GSE53757 datasets (P<0.05).
Nomogram modeling
To assess whether the risk score was an independent prognostic factor, univariate Cox analysis was performed on the TCGA-KIRC dataset, incorporating clinical characteristics (age, gender, stage, grade), with factors showing HR ≠1 and P<0.05 selected for multivariate Cox analysis to identify independent prognostic factors (HR ≠1 and P<0.05). A nomogram was then constructed to predict patient survival probability, with its performance validated by calibration and ROC curves. Additionally, Gene Set Enrichment Analysis (GSEA) was employed to explore the biological functions of the prognostic genes (background gene set: c2.cp.kegg.v7.4.symbols.gmt). Gene Set Variation Analysis (GSVA) was performed using the GSVA package (version 1.44.5) (27) with the “h.all.v2023.1.Hs.symbols” gene set from the Molecular Signatures Database (MSigDB) (https://www.gsea-msigdb.org/gsea/msigdb/index.jsp) to investigate differences in pathway enrichment between the high- and low-risk groups. Finally, ingenuity pathway analysis (IPA) was carried out through the QIAGEN IPA database (www.qiagen.com/ingenuity) to further elucidate the molecular mechanisms underlying the prognostic genes.
Tumor microenvironment (TME) analysis
To further explore the immune cell infiltration differences between high- and low-risk cohorts, the TME landscape was assessed. The CIBERSORT algorithm (28) was used to estimate the abundance of 22 immune cell types in each ccRCC sample, and infiltration proportions were compared between high- and low-risk groups. Spearman correlation analysis was then conducted to examine the relationships between immune cells and between immune cells and prognostic genes. Additionally, immune scores, stromal scores, and ESTIMATE scores were calculated for both risk cohorts and compared using the Wilcoxon test. Spearman correlation analysis was further performed to assess the relationship between these scores and prognostic genes.
Construction of a lncRNA-miRNA-mRNA and drug-gene networks
To identify upstream regulatory molecules of prognostic genes, a lncRNA-miRNA-mRNA network was constructed. Differentially expressed miRNAs (DE-miRNAs) and DE-lncRNAs between ccRCC and control cohorts in the TCGA-KIRC dataset were identified using the DESeq2 package (version 1.36.0) (25) with criteria |log2FC| >1 and adjusted P<0.05. Prognostic miRNAs were predicted using the miRTarBase database (https://mirtarbase.cuhk.edu.cn/), and the intersecting miRNAs were identified by comparing them with DE-miRNAs. lncRNAs associated with these intersecting miRNAs were retrieved from the starBase database (https://starbase.sysu.edu.cn/starbase2/), and the overlap with DE-lncRNAs led to the identification of intersected lncRNAs. Finally, the lncRNA-miRNA-mRNA network was constructed. Furthermore, the Drug-Gene Interaction Database (DGIdb) (https://www.dgidb.org/) was utilized to predict targeted therapeutic agents for the prognostic genes, and a drug-gene interaction network was constructed.
Quantitative real-time polymerase chain reaction (RT-qPCR)
Additionally, five pairs of ccRCC and para-cancer tissues were collected from patients with ccRCC who underwent surgery at The First Affiliated Hospital of Soochow University in China between February and June 2023. The tissue samples were diagnosed by two pathologists from the Department of Pathology at the same hospital. Ethical approval was granted by the Ethics Committee of The First Affiliated Hospital of Soochow University (approval No. 2023 Research Batch No. 476), and informed consent was obtained from each patient. This study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Immediately following surgery, the ccRCC and paired para-cancer tissues were stored in liquid nitrogen. Total RNA was extracted using a Trizol reagent (Beyotime, China) according to the manufacturer’s instructions. The quality and quantity of RNA were assessed using the Nanodrop ND-2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA), with RNA purity standards set between 1.8–2.1 for A260/A280. RNA concentrations were normalized using RNase-free ddH2O, and cDNA was synthesized using PrimeScript™ RT Master Mix (Takara Bio INC, Beijing, China). The cDNA samples were stored at −20 °C prior to use. RT-qPCR was performed using ChamQ SYBR qPCR Master Mix (Vazyme, Nanjing, China) and specific primers, following the steps: initial denaturation for 3 minutes at 95 °C, 15 seconds of denaturation at 95 °C, annealing, and extension for 40 seconds at 60 °C, with a total of 40 cycles. β-actin was used as a control to calculate the relative expression of target genes in ccRCC and para-cancer samples using the 2−ΔΔCt method (29). The primer sequences for model genes and β-actin were synthesized by Tsingke Biotechnology (Beijing, China), as detailed in Table 1.
Table 1
| Gene symbol | Primer sequence |
|---|---|
| β-actin | Forward primer: ACAGAGCCTCGCCTTTGCC |
| Reverse primer: GATATCATCATCCATGGTGAGCTGG | |
| PROS1 | Forward primer: TCTCAGAGGCAAACTTTTTGTCA |
| Reverse primer: AGAATTTGCACGACGCTTCC | |
| BMP6 | Forward primer: GCTCAACCGCAAGAGCCTTC |
| Reverse primer: TGTCGTACTCCACCAGGTTC | |
| RUNX1 | Forward primer: CATCGCTTTCAAGGTGGTGG |
| Reverse primer: ATGGCTGCGGTAGCATTTCT | |
| TIMP1 | Forward primer: ATTCCGACCTCGTCATCAGG |
| Reverse primer: GGACCTGTGGAAGTATCCGC |
RT-qPCR, quantitative real-time polymerase chain reaction.
Statistical analysis
Bioinformatic analyses were conducted in the R program (version 4.2). The results were regarded as significant at P<0.05.
Results
Definiteness of DEGs and the key module
A total of 3,311 DEGs were identified between ccRCC and control samples, with the heatmap illustrating the expression patterns of the top 20 upregulated and downregulated genes (Figure 1A, available online: https://cdn.amegroups.cn/static/public/tau-24-483-1.xlsx). These DEGs were associated with GO terms such as negative regulation of immune system processes, regulation of immune effector processes, leukocyte-mediated immunity, leukocyte activation in immune responses, and cell activation in immune responses (Figure 1B). Additionally, they were enriched in KEGG pathways, including glycine, serine, and threonine metabolism, as well as valine, leucine, and isoleucine degradation (Figure 1B). Figure 1C highlights two outlier samples in TCGA-KIRC (TCGA-B4-5832-01A, TCGA-CZ-5989-01A). Upon setting the soft threshold to 13 (R2=0.85), the network closely approximated a scale-free distribution with a shallow slope (Figure 1D). Ten modules were identified (Figure 1E), with the blue module, containing 1,719 genes, showing the strongest correlation with clinical traits (Cor =−0.82, P<0.001) and thus considered the key module (Figure 1F).

The DE-CFRGs were correlated with immune, and complement and coagulation cascade-related pathways
The intersection of DEGs, CFRGs, and key module genes yielded 16 DE-CFRGs (Figure 2A). These DE-CFRGs were enriched in GO terms such as negative regulation of cytokine production involved in immune system processes, immune responses, leukocyte activation in immune responses, regulation of immune effector processes, and cell activation in immune responses (Figure 2B). KEGG pathway analysis revealed their involvement in complement and coagulation cascades, Th17 cell differentiation, HIF-1 signaling, AGE-RAGE signaling in diabetic complications, and the Hippo signaling pathway (Figure 2C).

The prognostic model showed an excellent ability to predict the prognosis of patients with ccRCC
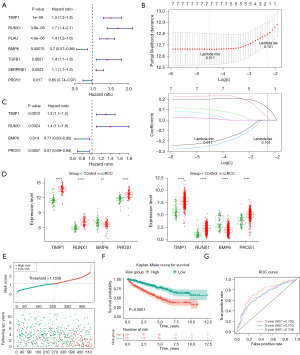
Four prognostic genes were identified: TIMP1, RUNX1, BMP6, and PROS1 (Figure 3A-3C), with consistent expression trends observed across both the TCGA-KIRC and GSE53757 datasets (Figure 3D). As shown in Figure 3E, survival samples predominantly clustered in the lower risk score region, while death samples were concentrated in the higher risk score region. K-M survival analysis demonstrated significantly shorter survival in the high-risk group compared to the low-risk group (Figure 3F). The prognostic model exhibited excellent predictive ability, with area under the curve (AUC) values exceeding 0.7 at 1 (AUC =0.750), 3 (AUC =0.701), and 5 years (AUC =0.704) (Figure 3G). Validation in the E-MTAB-1980 cohort confirmed these results (Figure S1).
The stage, risk score, and grade were correlated with ccRCC prognosis
The heatmap demonstrated significant differences in gender, grade, and stage between high- and low-risk cohorts (Figure 4A). Univariate and multivariate Cox regression analyses identified three independent prognostic factors: stage, risk score, and grade (Figure 4B). A nomogram was constructed, indicating a strong predictive ability for ccRCC, further validated by calibration and ROC curves (Figure 4C).

The prognostic model was related to immune, metabolism, and complement and coagulation cascade-related pathways
To explore the biological functions and pathways associated with the prognostic genes, GSEA was performed. TIMP1 was enriched in pathways related to intestinal immune network for immunoglobulin A (IgA) production, autoimmune thyroid disease, the citrate cycle [tricarboxylic acid (TCA) cycle], primary immunodeficiency, and valine, leucine, and isoleucine degradation. RUNX1 was involved in pathways including complement and coagulation cascades, pyruvate metabolism, butanoate metabolism, hematopoietic cell lineage, and neuroactive ligand-receptor interactions. BMP6 participated in WNT signaling, oxidative phosphorylation, MAPK signaling, neurotrophin signaling, and ECM receptor interactions. PROS1 was enriched in pathways such as arginine and proline metabolism, histidine metabolism, tryptophan metabolism, cysteine and methionine metabolism, and the renin-angiotensin system (Figure 5A, available online: https://cdn.amegroups.cn/static/public/tau-24-483-2.xlsx). Additionally, hallmark pathways such as inflammatory response, epithelial-mesenchymal transition, and interferon-gamma response were upregulated in both high- and low-risk groups, while bile acid metabolism, protein secretion, and oxidative phosphorylation were downregulated (Figure 5B). Further, prognostic genes were associated with classical pathways, including CREB signaling in neurons and the extrinsic prothrombin activation pathway (Figure 5C). An upstream and downstream regulatory network of prognostic genes was constructed, highlighting interactions such as ADIPOQ-ERK1/2 and RUNX1-Mlc (Figure 5D).

The prognostic model played a pivotal role in the immune microenvironment of ccRCC
The bar chart illustrates the distribution of 22 immune cell types across the high- and low-risk cohorts (Figure 6A). Twelve immune cell types exhibited significant differences in infiltration levels between the two groups, including resting memory CD4 T cells and plasma cells (Figure 6B). CD8 T cells showed the strongest positive correlation with helper follicular T cells (Cor =0.58, P<0.001) and the strongest negative correlation with resting memory CD4 T cells (Cor =−0.61, P<0.001) (Figure 6C). BMP6 was most positively correlated with resting mast cells (Cor =0.35, P<0.001) and most negatively correlated with regulatory T cells (Tregs) (Cor =−0.34, P<0.001) (Figure 6D). Immune and stromal cell infiltration was notably higher in the high-risk group, as indicated by elevated stromal, immune, and ESTIMATE scores compared to the low-risk group (Figure 6E). Scatter plots revealed that RUNX1 exhibited the highest correlations with immune score (Cor =0.36, P<0.001), stromal score (Cor =0.52, P<0.001), and ESTIMATE score (Cor =0.48, P<0.001) (Figure S2).

The lncRNA-miRNA-mRNA and drug-gene networks
A total of 257 DE-miRNAs and 983 DE-lncRNAs were identified between ccRCC and control samples in TCGA-KIRC (Figure 7A,7B). Four intersecting miRNAs and 33 intersecting lncRNAs were also obtained, resulting in a lncRNA-miRNA-mRNA network comprising 39 nodes and 39 edges. Notable interactions within the network included HOTAIR-hsa-miR-642a-5p-RUNX1, LUCAT1-hsa-miR-181a-5p-TIMP1, and CASC19-hsa-miR-181a-5p-TIMP1, among others (Figure 7C). Furthermore, 14 potential therapeutic agents for prognostic genes were predicted, and a drug-gene network was constructed, featuring interactions such as tamoxifen citrate-RUNX1, cytarabine-RUNX1, and menadione-PROS1 (Figure 7D). The chemical structures of these 14 therapeutic drugs are shown in Figure 7E.

Verification of TIMP1, RUNX1, BMP6, and PROS1 expression by RT-qPCR
RT-qPCR analysis was conducted to assess the expression levels of TIMP1, RUNX1, BMP6, and PROS1 in ccRCC and adjacent normal tissues. As depicted in Figure 8, all four genes were upregulated in cancer tissues, with TIMP1 and RUNX1 showing expression patterns consistent with bioinformatics analysis. However, the expression levels of BMP6 and PROS1 could not be validated in the available samples.

Discussion
Key findings
A novel prognostic model incorporating TIMP1, RUNX1, BMP6, and PROS1 was established for ccRCC, significantly enhancing prognostic prediction accuracy and revealing potential therapeutic targets and drugs. This model provides new insights into the roles of CFRGs in ccRCC progression.
Strengths and limitations
In this study, four key prognostic genes (PROS1, BMP6, RUNX1, TIMP1) associated with both coagulation and fibrinolysis in patients with ccRCC were identified, and a prognostic model was developed for the first time. While previous studies have focused on genes linked to the coagulation system in ccRCC, our approach uniquely considers the interconnectedness of the coagulation and fibrinolysis systems, emphasizing their dynamic equilibrium (13,30). Notably, our exploration of the PROS1 gene’s role in ccRCC is the first of its kind, providing new insight into its potential significance in this context. Additionally, RT-qPCR validation confirmed the expression of TIMP1 and RUNX1 in line with bioinformatics findings, though validation of BMP6 and PROS1 encountered challenges, possibly due to individual differences. This study offers promising avenues for further elucidating the pathogenesis of renal clear cell carcinoma from the perspective of coagulation and fibrinolysis and may guide subsequent research directions. Furthermore, it presents the possibility of discovering novel therapeutic targets for ccRCC and identifying more effective drugs for clinical use based on these new targets.
Comparison with similar research
Currently, survival rates for advanced ccRCC and metastatic ccRCC remain low, due to the limited efficacy of conventional therapies like chemotherapy and radiotherapy, with only a small proportion of patients benefiting from immunotherapy and targeted therapies (31). Available targeted therapies for advanced ccRCC are still limited, with most studies focusing on tyrosine kinase inhibitors, while other underlying mechanisms remain unexplored (32). Therefore, the identification of new therapeutic target candidates may pave the way for advancements in clinical strategies for ccRCC treatment (33).
Components of the coagulation and fibrinolysis systems play a critical role in carcinogenesis, cancer progression, and metastasis. For instance, in pancreatic cancer, increased clot-prone tumor cells are associated with enhanced platelet adhesion, while fibrin promotes tumor metastasis (24). In ccRCC, the presence of tumor thrombus in the inferior vena cava is often indicative of poor prognosis (34). Additionally, RCC metastasis may be linked to the activation of the blood clotting system through neutrophil extracellular traps (NETs), which are composed of intravascular neutrophils (35). Given these observations, it is hypothesized that CFRGs are clinically significant in ccRCC, motivating further investigation into their potential role in this malignancy.
Explanations of findings
Four CFRGs were identified in patients with ccRCC: TIMP1, RUNX1, BMP6, and PROS1. TIMP1, which inhibits matrix metalloproteinase (MMP) activity, modulates the degradation and remodeling of extracellular matrix components (36). Elevated TIMP1 levels have been linked to metastasis in ccRCC, suggesting its potential as a prognostic marker (37). RUNX1, a key regulator of hematopoiesis, is implicated in several malignancies, with higher expression correlating with poor prognosis in ccRCC (38,39). BMP6, a tumor suppressor gene, reduces invasion by promoting adhesion and intercellular interactions and inducing angiogenesis in endothelial cells (40). It also plays diverse roles in various cancers, including prostate cancer (41), malignant pleural mesothelioma (MPM) (42), and breast cancer (43). In RCC, BMP6 promotes tumor cell proliferation through interleukin 10 (IL-10)-mediated tumor-associated macrophage (TAM) M2 polarization (44). Additionally, BMP6 serves as a prognostic marker in cervical cancer, with its inclusion in a risk score model improving survival prediction for patients with squamous cell carcinoma (45). PROS1, a natural ligand for the TAM receptors Tyro3, Axl, and MerTK, suppresses M1 macrophage cytokine production in both in vitro and in vivo models, thereby enhancing tumor cell survival (46). Elevated PROS1 expression is associated with the progression of papillary thyroid carcinoma (47) and oral squamous cell carcinoma (48), though its role in RCC remains underexplored. These findings highlight the potential significance of these genes in ccRCC progression and prognosis, warranting further validation within risk assessment models.
GSEA and GSVA enrichment analyses revealed two key pathways strongly correlated with the prognostic model: the IL6/JAK/STAT3 pathway and fatty acid metabolism. STAT3, a member of the signal transducer and activator of transcription (STAT) family, interacts extensively with numerous proteins, serving as a key mediator between various oncogenes and offering substantial potential for targeted therapies (49). Studies have demonstrated that ccRCC-derived exosomes can promote tumorigenesis via the IL6/JAK/STAT3 pathway (50,51). Furthermore, increasing evidence indicates that fatty acid metabolism plays a significant role in ccRCC development (52), with lipid metabolism disorders commonly observed in ccRCC (53). Tumor cells in ccRCC tissues may enhance fatty acid uptake through CD36 and FATP4-mediated transmembrane transport (54). Notably, targeting fatty acid metabolism could alleviate pain associated with ccRCC (55). While these pathways are well-established in cancer research, their specific connection to ccRCC requires further investigation.
Three independent prognostic factors were identified in this study: staging, risk score, and grading. The grading of RCC, including the Fuhrman grading system and the World Health Organization/International Society of Urological Pathology (WHO/ISUP) classification, has long been recognized as a prognostic indicator (56). Regarding staging, advanced stages significantly correlate with reduced survival rates in ccRCC, a finding confirmed by our analysis (57). K-M plots in this study demonstrated the robust predictive ability of the prognostic model for ccRCC, further validated by calibration curves and ROC curves.
The role of immune cells in the TME is also pivotal. Current literature suggests that an increased abundance of Tregs and follicular helper T cells (Tfh) is associated with poorer outcomes in patients with ccRCC, whereas a higher presence of resting mast cells correlates with more favorable prognoses (58,59). Notably, the accumulation of Tregs in the TME may correlate with a heightened angiogenic state in RCC (60). In the prognostic model, BMP6 acted as a protective factor for ccRCC, with immunoassay results indicating that BMP6 was positively correlated with resting mast cells and negatively correlated with Tregs. Conversely, TIMP1, identified as a risk factor for ccRCC, showed a positive correlation with Tregs. These findings align with previous studies, supporting the validity of our results. Additionally, TIMP1 is closely linked to immune checkpoints such as CTLA4 and CD96, suggesting potential therapeutic targets (37).
This study also predicted potential therapeutic agents for the four prognostic genes and constructed drug-gene networks, including disulfiram-RUNX1, estradiol-PROS1, progesterone-PROS1, and other related pairs. Disulfiram has been shown to inhibit various cancers by targeting the ubiquitin-proteasome system via NPL4 inhibition, and it synergizes with Sunitinib to reduce renal cancer cell proliferation (61). RCC is responsive to sex hormones, with downregulation of progesterone receptor-related genes correlating with poor tumor prognosis (62). Estradiol can induce growth inhibition in RCC through estrogen receptor (ER) activation (63). These drug mechanisms warrant further validation through cell and animal experiments. While existing studies support the feasibility of these therapies, it is anticipated that as the mechanisms underlying the fibrinolytic-coagulation system in renal cancer become clearer in the next 5 years, new drugs will emerge, progressing from animal models to clinical trials and ultimately improving patient survival rates.
Implications and actions needed
This study has several limitations. First, only the expression levels of selected genes were verified, and the small sample size must be expanded in future studies. Additionally, RNA-level validation alone does not fully capture cellular function, and the role of individual genes in the proliferation, invasion, and migration of renal cancer cells should be further explored at the protein expression level. Animal studies are also needed to validate the function of these genes in vivo, thereby strengthening the evidence. Furthermore, this study relied on TCGA database analysis, and future research will incorporate multi-center data and clinical trials. In conclusion, we would continue to take care of the research progress of the coagulation and fibrinolytic system and ccRCC, in expecting to have more treatment options based on this study.
Conclusions
In conclusion, a prognostic model linking genes associated with the coagulation and fibrinolysis systems to ccRCC was established through bioinformatics analysis and basic experimental validation, contributing insights into the pathogenesis of renal clear cell carcinoma and offering potential therapeutic avenues. However, several limitations should be noted. While the prognostic model demonstrated promising results, its clinical applicability needs further validation through prospective clinical studies and real-world testing before it can be widely implemented in clinical practice.
Acknowledgments
The authors express their gratitude to the entire team for their invaluable support and assistance throughout the course of this study.
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://tau.amegroups.com/article/view/10.21037/tau-24-483/rc
Data Sharing Statement: Available at https://tau.amegroups.com/article/view/10.21037/tau-24-483/dss
Peer Review File: Available at https://tau.amegroups.com/article/view/10.21037/tau-24-483/prf
Funding: This work was supported by
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tau.amegroups.com/article/view/10.21037/tau-24-483/coif). The authors declare no conflicts of interest.
Ethical Statement: The authors are accountable for all aspects of the work, ensuring that any concerns related to the accuracy or integrity of any part of the work are appropriately addressed. This study was conducted in accordance with the Declaration of Helsinki (as revised in 2013) and was approved by the Ethics Committee of The First Affiliated Hospital of Soochow University (approval No. 2023 Research Batch No. 476). Informed consent was obtained from all participants.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Laversanne M, Sung H, et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2024;74:229-63. [Crossref] [PubMed]
- Hsieh JJ, Purdue MP, Signoretti S, et al. Renal cell carcinoma. Nat Rev Dis Primers 2017;3:17009. [Crossref] [PubMed]
- Kathuria-Prakash N, Drolen C, Hannigan CA, et al. Immunotherapy and Metastatic Renal Cell Carcinoma: A Review of New Treatment Approaches. Life (Basel) 2021;12:24. [Crossref] [PubMed]
- Atkins MB, Tannir NM. Current and emerging therapies for first-line treatment of metastatic clear cell renal cell carcinoma. Cancer Treat Rev 2018;70:127-37. [Crossref] [PubMed]
- Jonasch E, Gao J, Rathmell WK. Renal cell carcinoma. BMJ 2014;349:g4797. [Crossref] [PubMed]
- Guven DC, Erul E, Kaygusuz Y, et al. Immune checkpoint inhibitor-related hearing loss: a systematic review and analysis of individual patient data. Support Care Cancer 2023;31:624. [Crossref] [PubMed]
- Rizzo A, Santoni M, Mollica V, et al. Peripheral neuropathy and headache in cancer patients treated with immunotherapy and immuno-oncology combinations: the MOUSEION-02 study. Expert Opin Drug Metab Toxicol 2021;17:1455-66. [Crossref] [PubMed]
- Choueiri TK, Kaelin WG Jr. Targeting the HIF2-VEGF axis in renal cell carcinoma. Nat Med 2020;26:1519-30. [Crossref] [PubMed]
- Mollica V, Santoni M, Matrana MR, et al. Concomitant Proton Pump Inhibitors and Outcome of Patients Treated with Nivolumab Alone or Plus Ipilimumab for Advanced Renal Cell Carcinoma. Target Oncol 2022;17:61-8. [Crossref] [PubMed]
- Michaelis J, Grabbert M, Sigle A, et al. Tyrosine Kinase Inhibitors in the Treatment of Metastasised Renal Cell Carcinoma-Future or the Past? Cancers (Basel) 2022;14:3777. [Crossref] [PubMed]
- Rosellini M, Marchetti A, Mollica V, et al. Prognostic and predictive biomarkers for immunotherapy in advanced renal cell carcinoma. Nat Rev Urol 2023;20:133-57. [Crossref] [PubMed]
- Sahin TK, Rizzo A, Aksoy S, et al. Prognostic Significance of the Royal Marsden Hospital (RMH) Score in Patients with Cancer: A Systematic Review and Meta-Analysis. Cancers (Basel) 2024;16:1835. [Crossref] [PubMed]
- Fu Y, Liu Y, Jin Y, et al. Value of Coagulation and Fibrinolysis Biomarker in Lung Cancer Patients with Thromboembolism. Zhongguo Fei Ai Za Zhi 2018;21:583-87. [PubMed]
- McMahon BJ, Kwaan HC. Components of the Plasminogen-Plasmin System as Biologic Markers for Cancer. Adv Exp Med Biol 2015;867:145-56. [Crossref] [PubMed]
- Urban P, Vuaroqueaux V, Labuhn M, et al. Increased expression of urokinase-type plasminogen activator mRNA determines adverse prognosis in ErbB2-positive primary breast cancer. J Clin Oncol 2006;24:4245-53. [Crossref] [PubMed]
- Halamkova J, Kiss I, Pavlovsky Z, et al. Clinical significance of the plasminogen activator system in relation to grade of tumor and treatment response in colorectal carcinoma patients. Neoplasma 2011;58:377-85. [Crossref] [PubMed]
- Yang SF, Hsieh YS, Lin CL, et al. Increased plasma levels of urokinase plasminogen activator and matrix metalloproteinase-9 in nonsmall cell lung cancer patients. Clin Chim Acta 2005;354:91-9. [Crossref] [PubMed]
- Rubina KA, Sysoeva VY, Zagorujko EI, et al. Increased expression of uPA, uPAR, and PAI-1 in psoriatic skin and in basal cell carcinomas. Arch Dermatol Res 2017;309:433-42. [Crossref] [PubMed]
- Taubert H, Würl P, Greither T, et al. Co-detection of members of the urokinase plasminogen activator system in tumour tissue and serum correlates with a poor prognosis for soft-tissue sarcoma patients. Br J Cancer 2010;102:731-7. [Crossref] [PubMed]
- Liu Q, Li W, Yang S, et al. High expression of uPA related to p38MAPK in esophageal cancer indicates poor prognosis. Onco Targets Ther 2018;11:8427-34. [Crossref] [PubMed]
- Ma J, Qi G, Xu J, et al. Overexpression of forkhead box M1 and urokinase-type plasminogen activator in gastric cancer is associated with cancer progression and poor prognosis. Oncol Lett 2017;14:7288-96. [Crossref] [PubMed]
- Repetto O, De Re V. Coagulation and fibrinolysis in gastric cancer. Ann N Y Acad Sci 2017;1404:27-48. [Crossref] [PubMed]
- Lal I, Dittus K, Holmes CE. Platelets, coagulation and fibrinolysis in breast cancer progression. Breast Cancer Res 2013;15:207. [Crossref] [PubMed]
- Fang L, Xu Q, Qian J, et al. Aberrant Factors of Fibrinolysis and Coagulation in Pancreatic Cancer. Onco Targets Ther 2021;14:53-65. [Crossref] [PubMed]
- Liu S, Wang Z, Zhu R, et al. Three Differential Expression Analysis Methods for RNA Sequencing: limma, EdgeR, DESeq2. J Vis Exp 2021; [Crossref] [PubMed]
- Yu G, Wang LG, Han Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 2012;16:284-7. [Crossref] [PubMed]
- Su R, Jin C, Bu H, et al. Development and Validation of an Immune-Related Prognostic Signature in Cervical Cancer. Front Oncol 2022;12:861392. [Crossref] [PubMed]
- Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods 2015;12:453-7. [Crossref] [PubMed]
- Sninsky JA, Liu S, Eluri S, et al. CSTB and FABP5 Serum mRNA Differentiate Histologically Active and Inactive Patients With Eosinophilic Esophagitis. Gastro Hep Adv 2023;3:1-3. [Crossref] [PubMed]
- Yin G, Tian T, Ji X, et al. Integrated analysis to identify the prognostic and immunotherapeutic roles of coagulation-associated gene signature in clear cell renal cell carcinoma. Front Immunol 2023;14:1107419. [Crossref] [PubMed]
- Cheng G, Li M, Ma X, et al. Systematic Analysis of microRNA Biomarkers for Diagnosis, Prognosis, and Therapy in Patients With Clear Cell Renal Cell Carcinoma. Front Oncol 2020;10:543817. [Crossref] [PubMed]
- Yang S, Yang X, Hou Z, et al. Rationale for immune checkpoint inhibitors plus targeted therapy for advanced renal cell carcinoma. Heliyon 2024;10:e29215. [Crossref] [PubMed]
- Ajadee A, Mahmud S, Hossain MB, et al. Screening of differential gene expression patterns through survival analysis for diagnosis, prognosis and therapies of clear cell renal cell carcinoma. PLoS One 2024;19:e0310843. [Crossref] [PubMed]
- Ljungberg B, Bensalah K, Canfield S, et al. EAU guidelines on renal cell carcinoma: 2014 update. Eur Urol 2015;67:913-24. [Crossref] [PubMed]
- Wen L, Guo L, Zhang W, et al. Cooperation Between the Inflammation and Coagulation Systems Promotes the Survival of Circulating Tumor Cells in Renal Cell Carcinoma Patients. Front Oncol 2019;9:504. [Crossref] [PubMed]
- Batra J, Robinson J, Soares AS, et al. Matrix metalloproteinase-10 (MMP-10) interaction with tissue inhibitors of metalloproteinases TIMP-1 and TIMP-2: binding studies and crystal structure. J Biol Chem 2012;287:15935-46. [Crossref] [PubMed]
- Li Q, Wei K, Zhang X, et al. TIMP1 shapes an immunosuppressive microenvironment by regulating anoikis to promote the progression of clear cell renal cell carcinoma. Aging (Albany NY) 2023;15:8908-29. [Crossref] [PubMed]
- Yzaguirre AD, de Bruijn MF, Speck NA. The Role of Runx1 in Embryonic Blood Cell Formation. Adv Exp Med Biol 2017;962:47-64. [Crossref] [PubMed]
- Fu Y, Sun S, Man X, et al. Increased expression of RUNX1 in clear cell renal cell carcinoma predicts poor prognosis. PeerJ 2019;7:e7854. [Crossref] [PubMed]
- García Muro AM, García Ruvalcaba A, Rizo de la Torre LDC, et al. Role of the BMP6 protein in breast cancer and other types of cancer. Growth Factors 2021;39:1-13. [Crossref] [PubMed]
- Dai J, Keller J, Zhang J, et al. Bone morphogenetic protein-6 promotes osteoblastic prostate cancer bone metastases through a dual mechanism. Cancer Res 2005;65:8274-85. [Crossref] [PubMed]
- Kimura K, Toyooka S, Tsukuda K, et al. The aberrant promoter methylation of BMP3b and BMP6 in malignant pleural mesotheliomas. Oncol Rep 2008;20:1265-8. [PubMed]
- Wang C, Hu F, Guo S, et al. BMP-6 inhibits MMP-9 expression by regulating heme oxygenase-1 in MCF-7 breast cancer cells. J Cancer Res Clin Oncol 2011;137:985-95. [Crossref] [PubMed]
- Lee JH, Lee GT, Woo SH, et al. BMP-6 in renal cell carcinoma promotes tumor proliferation through IL-10-dependent M2 polarization of tumor-associated macrophages. Cancer Res 2013;73:3604-14. [Crossref] [PubMed]
- Wang Q, Vattai A, Vilsmaier T, et al. Immunogenomic Identification for Predicting the Prognosis of Cervical Cancer Patients. Int J Mol Sci 2021;22:2442. [Crossref] [PubMed]
- Ubil E, Caskey L, Holtzhausen A, et al. Tumor-secreted Pros1 inhibits macrophage M1 polarization to reduce antitumor immune response. J Clin Invest 2018;128:2356-69. [Crossref] [PubMed]
- Wang J, Lei M, Xu Z. Aberrant expression of PROS1 correlates with human papillary thyroid cancer progression. PeerJ 2021;9:e11813. [Crossref] [PubMed]
- Abboud-Jarrous G, Priya S, Maimon A, et al. Protein S drives oral squamous cell carcinoma tumorigenicity through regulation of AXL. Oncotarget 2017;8:13986-4002. [Crossref] [PubMed]
- Erdogan F, Radu TB, Orlova A, et al. JAK-STAT core cancer pathway: An integrative cancer interactome analysis. J Cell Mol Med 2022;26:2049-62. [Crossref] [PubMed]
- Zhang W, Zheng X, Yu Y, et al. Renal cell carcinoma-derived exosomes deliver lncARSR to induce macrophage polarization and promote tumor progression via STAT3 pathway. Int J Biol Sci 2022;18:3209-22. [Crossref] [PubMed]
- Shen T, Miao S, Zhou Y, et al. Exosomal AP000439.2 from clear cell renal cell carcinoma induces M2 macrophage polarization to promote tumor progression through activation of STAT3. Cell Commun Signal 2022;20:152. [Crossref] [PubMed]
- Zhao Z, Liu Y, Liu Q, et al. The mRNA Expression Signature and Prognostic Analysis of Multiple Fatty Acid Metabolic Enzymes in Clear Cell Renal Cell Carcinoma. J Cancer 2019;10:6599-607. [Crossref] [PubMed]
- Heravi G, Yazdanpanah O, Podgorski I, et al. Lipid metabolism reprogramming in renal cell carcinoma. Cancer Metastasis Rev 2022;41:17-31. [Crossref] [PubMed]
- Kim YS, Jung J, Jeong H, et al. High Membranous Expression of Fatty Acid Transport Protein 4 Is Associated with Tumorigenesis and Tumor Progression in Clear Cell Renal Cell Carcinoma. Dis Markers 2019;2019:5702026. [Crossref] [PubMed]
- Ding R, Wei H, Jiang X, et al. Prognosis and pain dissection of novel signatures in kidney renal clear cell carcinoma based on fatty acid metabolism-related genes. Front Oncol 2022;12:1094657. [Crossref] [PubMed]
- Delahunt B, Eble JN, Egevad L, et al. Grading of renal cell carcinoma. Histopathology 2019;74:4-17. [Crossref] [PubMed]
- Erman M, Benekli M, Basaran M, et al. Renal cell cancer: overview of the current therapeutic landscape. Expert Rev Anticancer Ther 2016;16:955-68. [Crossref] [PubMed]
- Zhu G, Pei L, Yin H, et al. Profiles of tumor-infiltrating immune cells in renal cell carcinoma and their clinical implications. Oncol Lett 2019;18:5235-42. [Crossref] [PubMed]
- Wang Q, Tang H, Luo X, et al. Immune-Associated Gene Signatures Serve as a Promising Biomarker of Immunotherapeutic Prognosis for Renal Clear Cell Carcinoma. Front Immunol 2022;13:890150. [Crossref] [PubMed]
- Ning H, Shao QQ, Ding KJ, et al. Tumor-infiltrating regulatory T cells are positively correlated with angiogenic status in renal cell carcinoma. Chin Med J (Engl) 2012;125:2120-5. [PubMed]
- Yoshino H, Yamada Y, Enokida H, et al. Targeting NPL4 via drug repositioning using disulfiram for the treatment of clear cell renal cell carcinoma. PLoS One 2020;15:e0236119. [Crossref] [PubMed]
- Tao C, Liu W, Yan X, et al. PAQR5 Expression Is Suppressed by TGFβ1 and Associated With a Poor Survival Outcome in Renal Clear Cell Carcinoma. Front Oncol 2022;11:827344. [Crossref] [PubMed]
- Chen KC, Lin CM, Huang CJ, et al. Dual Roles of 17-β Estradiol in Estrogen Receptor-dependent Growth Inhibition in Renal Cell Carcinoma. Cancer Genomics Proteomics 2016;13:219-30. [PubMed]


