
Piperlongumine inhibits renal cell carcinoma progression by modulating the DDX11-miR-15b-3p-DLD axis
Highlight box
Key findings
• The study discovered that piperlongumine (PL) can significantly inhibit the progression of renal cell carcinoma (RCC) by interfering with the mitochondrial homeostasis of RCC cells.
• PL-induced inhibition of RCC occurs through the modulation of the DDX11–miR-15b-3p-dihydrolipoamide dehydrogenase (DLD) axis.
What is known and what is new?
• PL is a natural alkaloid that can exert anticancer effects. However, research on the role of PL in RCC remains limited.
• This study is the first to report that PL significantly inhibits RCC proliferation. In terms of mechanism, PL reduces the expression of DDX11, inhibits the maturation of miR-15b-3p, and further elevates the level of DLD.
What is the implication, and what should change now?
• As a natural compound, PL has minimal side effects. Moreover, it can significantly inhibit the progression of RCC and may serve as a therapeutic target for this disease.
• Future research should concentrate on the impact of PL on RCC metastasis and clarify the exact mechanisms by which PL regulates RCC metastasis.
Introduction
Renal cell carcinoma (RCC), a fatal malignant tumor that originates from the urinary tubular epithelial system within the renal parenchyma, accounts for approximately 2% of all cancers (1). The subtypes of RCC include clear-cell RCC (ccRCC), papillary RCC (pRCC), and chromophobe RCC (chRCC), among which ccRCC is the most prevalent, accounting for 70–80% of all kidney cancers (2). The onset of RCC often lacks typical clinical symptoms, resulting in nearly 20% of initial diagnoses being cases of metastatic RCC (3,4). Although targeted therapy and immunotherapy can partly impede disease progression, the overall treatment effect remains unsatisfactory, with a 5-year survival rate of less than 20% (5). Therefore, clarifying the specific mechanisms of RCC occurrence and development may aid considerably in the prevention and treatment of RCC.
Piperlongumine (PL) is a natural alkaloid obtained from the long pepper (6). The chemical formula for PL is C17H19NO5, and its relative molecular mass is 317.34. PL plays an important role in various physiological processes, such as in the exertion of antidiabetic, antiaging, and antioxidant effects, along with antiplatelet aggregation (7,8). In recent years, numerous studies have demonstrated PL’s ability to treat various tumor types (6). However, research on PL’s effect in RCC remains limited, and thus a comprehensive investigation into the role of PL in RCC could enhance the understanding of its mechanisms of action.
The DEAD-box helicase (DDX) protein family belongs to the eukaryotic RNA helicase superfamily (9). DDX family proteins are involved in regulating various essential biological processes, including translation, alternative splicing, messenger RNA (mRNA) nuclear export, microRNA (miRNA) regulation, and RNA degradation (9,10). In recent years, numerous studies have highlighted the close association between the abnormal expression of DDX family proteins and the occurrence and development of tumors. For example, DDX3 can impede the progression of colorectal cancer by modulating the MAPK signaling pathway, thereby activating the E-cadherin and β-catenin pathways (11). However, there are no reports on the specific mechanism by which DDX11 influences the occurrence and development of renal cancer.
Ferroptosis is a newly discovered form of cell death that is present across a wide variety of species (12). It is an iron-dependent form of regulated cell death caused by uncontrolled lipid peroxidation on the cell membrane (13). Ferroptosis can be stimulated through both intrinsic or extrinsic pathways. The intrinsic pathway is activated by the blockade of intracellular antioxidant enzymes such as GPX4 and SLC7A11, while the extrinsic pathway is initiated by the inhibition of cell membrane transporters (14). Recent research has discovered the significance of ferroptosis in tumor biology and cancer therapy, especially in RCC (15). For example, one study found that DPP9 can stabilize NRF2, promote the transcription of SLC7A11, and further suppress ferroptosis in RCC cells (16). However, the specific mechanism by which PL influences ferroptosis remains unclear.
In this study, we found that PL could significantly inhibit the progression of RCC through interfering with the mitochondrial homeostasis of RCC cells. In terms of mechanism, our analysis indicated that PL decreased the expression of DDX11, inhibited the maturation of miR-15b-3p, and further increased the level of dihydrolipoamide dehydrogenase (DLD). Moreover, in vivo assays further demonstrated that DDX11 contributes significantly to the PL-induced inhibition of RCC. Taken together, our findings suggest that DDX11 plays a critical role in PL-induced inhibition of RCC by modulating the miR-15b-3p-DLD axis. We present this article in accordance with the ARRIVE reporting checklist (available at https://tau.amegroups.com/article/view/10.21037/tau-2025-11/rc).
Methods
Cells and culture conditions
Human RCC cell lines (786-O, 769-P, OSRC-2, ACHN, and Caki-1), normal renal epithelial cell line, and HK-2 were all obtained from the China Cell Bank (Shanghai, China). The RCC cell lines were cultured in RPMI-1640 medium (Gibco, Thermo Fisher Scientific, Waltham, MA, USA) containing 10% fetal bovine serum (Gibco) with 5% CO2 at 37 ℃.
Cell transfection
The DDX11 short hairpin RNA (shRNA) and overexpression plasmids were purchased from GeneChem (Shanghai, China). The miR-15b-3p inhibitors and mimics were designed and purchased from GenePharma (Shanghai, China). Lipofectamine 3000 (Invitrogen, Thermo Fisher Scientific) reagent was applied for DDX11 shRNA and overexpression plasmid transfection. RNAimax (Invitrogen) reagent was used for miR-15b-3p inhibitor and mimic transfection. The sequences of the DDX11 shRNA were as follows: DDX11_sh1, 5'-GATCGACAACATCAACCTGTT-3'; and DDX11_sh2, 5'-CGACTCCTTGAAACTGGAACT-3'.
Western blot analysis
The proteins were lysed with RIPA (Beyotime Biotechnology, Shanghai, China) reagent and then subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The proteins were separated via SDS-PAGE and then transferred to a polyvinylidene fluoride (PVDF) membrane (Bio-Rad Laboratories, Hercules, CA, USA). Following this, the proteins were incubated with the primary antibodies at 4 ℃ overnight. Subsequently, the membrane was treated with secondary antibodies. After three washes, the signaling was obtained using a chemiluminescence system (Bio-Rad Laboratories). DDX11 (1:1,000; sc-271711; RRID: AB_10707661) and β-actin (1:5,000; sc-8432; RRID: AB_626630) antibodies were purchased from Santa Cruz Biotechnology Inc (Dallas, TX, USA). DGCR8 (1:1,000; ab191875; RRID: AB_2892625) and Drosha (1:1,000; ab303544; AB_303544), DLD (1:1,000; ab5258-1; RRID: AB_10896675) and were purchased from Abcam (Cambridge, UK).
Reverse-transcription quantitative polymerase chain reaction (qPCR)
Total RNA was extracted with TRIzol reagent (Tiangen Biotech, Beijing, China) according to the manufacturer’s instruction. The RNA was then reverse transcribed to complement DNA (cDNA) using PrimeScript RT Master Mix (Takara Bio, Kusatsu, Japan). Furthermore, the relative mRNA expression was detected using qPCR with specific gene primers. The primer sequences were as follows: β-actin forward, 5'-TTGCCGACAGGATGCAGAAGGA-3'; β-actin reverse, 5'-AGGTGGACAGCGAGGCCAGGAT-3'; DDX11 forward 5'-TGGAACTGGCCCCTTACATGA-3'; and DDX11 reverse, 5'-CTGCACAAACTGAGTAACCCA-3'.
RNA immunoprecipitation (RIP) assays
The Magna RIP kit (Merck Millipore, Darmstadt, Germany) was used to conduct RIP experiments according to the manufacturer’s instructions. Briefly, RCC cells were lysed using lysis buffer and incubated with specific antibodies or IgG at room temperature for 30 min. The complex was then incubated with magnetic beads at 4 ℃ overnight. After three washed, the immune-precipitated RNA was extracted and quantified using reverse-transcription qPCR (RT-qPCR).
RNA sequencing
The RNA sequencing was conducted by Lianchuan Biotechnologies (Hangzhou, China). Briefly, total RNA was extracted from PL-treated and control 786-O cells. RNA quality control included quantification via a NanoDrop spectrophotometer (OD260/280 ratio: 1.8–2.2) and integrity assessment using the Agilent Bioanalyzer (RNA Integrity Number, RIN ≥7). For mRNA sequencing, polyadenylated RNA was enriched using oligo(dT) magnetic beads to isolate poly(A)+ transcripts. The enriched RNA was fragmented into 150–300 bp segments, followed by cDNA synthesis with random hexamer primers or oligo(dT) primers. Double-stranded cDNA was then subjected to end repair, 3'-end adenylation, and adapter ligation. Libraries were amplified via PCR and validated for size distribution (200–400 bp, Agilent Bioanalyzer) and molar concentration (Qubit assay). Sequencing was performed on an Illumina platform using 2×150 bp paired-end mode, generating 30–50 million reads per sample. Differential gene expression analysis was performed using DESeq2 or edgeR, with functional enrichment of significant genes analyzed via Gene Ontology (GO) terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways using clusterProfiler [Benjamini-Hochberg correction, false discovery rate (FDR) <0.05].
Cell Counting Kit-8 (CCK-8) assays and colony formation assays
For CCK-8, the wild-type or transfected RCC cells were seeded into 96-well plates at a density of 2,000 cells/well. Cell vitality was detected using CCK-8 reagent (Yeasen Biotechnology, Shanghai, China) at 96 h. The half maximal inhibitory concentration (IC50) of PL in RCC and normal HK-2 cells was determined through the application of 0, 2, 4, 6, or 8 µM PL in these cells for 48 h, after which the cell vitality was detected using CCK-8 reagent.
For colony formation assays, the wild-type or transfected RCC cells were seeded into six-well plates at a density of 1,000 cells/well. After 10–14 days, the colonies were fixed and stained with 0.1% crystal violet.
Cell cycle analysis
The wild-type RCC cells were cultured in a six-well plate. After RCC cell attachment, 2 or 5 µM of PL was added. After treatment for 48 h, the RCC cells were harvested and washed twice. The cells were then incubated with propidium iodide (Multi Sciences Biotech, Hangzhou, China) for 30 min in the dark. After two washes, the cells examined via cytometry.
Detection of cellular reactive oxygen species (ROS), Fe2+ level, and mitochondrial membrane potential (MMP)
For the ROS assay, the Reactive Oxygen Species Assay Kit (Beyotime Biotechnology) was applied for cellular ROS detection according to the manufacturer’s instruction. Briefly, DCFH-DA was incubated with cells for 20 min after PL treatment or transfected at 37 ℃. Following this, the cells were washed three times, and the fluorescence signals were detected via cytometry.
For the MMP assay, the MMP changes were measured with a MMP assay kit with JC-1 (Beyotime Biotechnology). Briefly, the PL-treated or transfected cells were incubated with JC-1 staining fluid and protected from light for 20 min. Subsequently, the cells were washed three times, and the fluorescence signals were detected via cytometry.
For the Fe2+ assay, an iron assay kit (Sigma-Aldrich, St. Louis, MI, USA) was applied to measure the Fe2+ level in each sample according to the manufacturer’s instruction.
Immunofluorescence
The RCC cells were seeded in 24-well plates overnight. The cells were then fixed with 0.2% Triton X-100 for 12 min. After being washed three times with phosphate-buffered saline (PBS), the cells were blocked with 5% bovine serum albumin (BSA) for 1 hour at room temperature, and then MitoTracker Red (Beyotime Biotechnology) was applied to stain the cells for 30 min in the dark. After three washes, the morphology of mitochondria was visualized under a fluorescence microscope.
Coimmunoprecipitation
RCC cells in a six-well plate at 90% confluence were lysed with 1 mL of 1% NP-40 lysis buffer (Beyotime coimmunoprecipitation). After centrifugation for 30 mins at 11,000 ×g at 4 ℃, the supernatant was collected and incubated with DDX11 or IgG antibodies overnight. Protein A/G agarose beads were then added to the lysate for 4 hours at 4 ℃. Finally, the bead-antibody-protein complex was washed five times with 1% NP-40, with gentle shaking for 5 min each time. Finally, 1× loading buffer was added to elute the target proteins, which was followed by Western blotting.
Xenograft mouse model
The experimental protocols were performed under a project license (No. SFY-2023Y009) granted by the Ethic Committee of the Affiliated Hospital of Shaoxing University, in compliance with national guidelines for the care and use of animals. Twenty 4-week-old male nude mice (species: BALB/C; qualification no. B231030195; breeding application No. SYXK2022-0022) were randomly divided into five groups (n=5 per group). Stably transfected 769-P cells were injected subcutaneously into the flank of the BALB/c nude mice. When the tumor volume reached ~150 mm3, 5 mg/kg of PL (Selleck Chemicals, Houston, TX, USA) was applied to mice via intraperitoneal injection three times per week. After 6 weeks, the mice were killed via cervical dislocation following the administration of pentobarbital anesthesia. The tumor volume was calculated with the following formula: tumor volume = 0.5 × length × width2. All methods were performed in accordance with relevant guidelines and regulations.
Immunohistochemistry (IHC)
For IHC, RCC tissue sample was first fixed using formaldehyde. Subsequently, the fixed tissue was embedded in paraffin and cut into 3- to 5-mm-thick sections. These sections were then mounted on glass slides. Next, antigen retrieval techniques were applied to the samples to unmask or restore the antigenic sites. Prior to incubation with the primary antibodies, the tissue sections were incubated with BSA to prevent nonspecific binding of the antibodies. After this blocking step, the primary antibody, at a concentration of 1:400, was added to the tissue sections and incubated at 4 ℃ overnight to allow it to bind to the antigen in the tissue.
The sections were then washed three times with PBS. Subsequently, the secondary antibodies were added and allowed to bind to the primary antibody for 60 min at room temperature.
Finally, diaminobenzidine (DAB) was used as the substrate, which reacted to produce a brown color. The IHC results were then photographed under a light microscope.
Statistical analysis
All experiments were performed independently at least three times. The statistical analysis was performed via GraphPad Prism software (Dotmatics, Boston, MA, USA). The values are presented as the mean ± standard error of the mean (SEM). The Student t-test was applied to analyze the differences between two groups. P<0.05 was considered as statistically significant.
Results
PL inhibited RCC cell proliferation
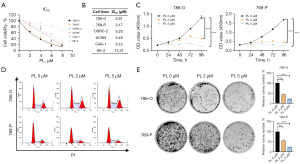
First, in order to examine the cytotoxic effect of PL on RCC cells, we treated different RCC cell lines (769-P, 786-O, ACHN, Caki-1, and OSRC-2) and a normal renal epithelial cell line, HK-2, with varying concentrations of PL. Surprisingly, we found that the RCC cell lines were more sensitive to PL than was HK-2. Furthermore, the 786-O and 769-P cells were more sensitive to PL treatment as compared to the other RCC cell lines, and the IC50 of PL in 786-O and 769-P cell lines was 2.81 and 3.17 µM, respectively (Figure 1A,1B). Therefore, we selected the 786-O and 769-P cell lines for further experiments. To characterize the broader response spectrum of PL, we selected two concentrations (2 and 5 µM) and comprehensively evaluated the dose-response relationship of PL with cell viability. Subsequently, we conducted CCK-8 assays after treating 786-O and 769-P cells with 2 and 5 µM of PL, revealing a significant inhibition of TCC cell proliferation, with the inhibition effect increasing with higher concentrations (Figure 1C). Cell cycle assays demonstrated that PL treatment arrested RCC cells in the G0–1 phase (Figure 1D). Additionally, colony formation assays indicated a significant decrease in the colony formation ability of RCC cells following PL treatment (Figure 1E).
PL impaired mitochondrial homeostasis
To further examine the specific effect of PL in impeding the progression of RCC, we subjected 786-O cells to a concentration of 2 µM PL for 48 h. Subsequently, both the treated and control groups underwent RNA-seq analysis. The results indicated that there were 762 differentially expressed genes (log2|fold change| >0.66 and false-discovery rate-adjusted P<0.05) after PL treatment (Figure 2A). We then performed KEGG analysis and observed that the dysregulated genes were enriched in mitochondrial-related pathways, particularly oxidative phosphorylation and ferroptosis (Figure 2B). Therefore, we hypothesized that PL may inhibit RCC cell proliferation by disrupting mitochondrial homeostasis. Subsequently, we measured the levels of ROS, MMP, and mitochondrial Fe2+ RCC cells treated with 2 and 5 µM of PL. The results indicated that PL significantly increased the levels of ROS and Fe2+ within the mitochondria, while reducing the MMP (Figure 2C-2E). We then performed immunofluorescence experiments using MitoTracker red on cells treated with PL and the control cells. The results revealed a fragmented mitochondrial morphology in RCC cells following PL treatment (Figure 2F). These data suggest that PL interferes with the mitochondrial homeostasis of RCC cells.

PL decreased DDX11 expression in RCC
To investigate how PL regulates mitochondrial homeostasis to impede the progression of RCC, we first analyzed the RNA-seq data. Based on the sequencing results, we selected DDX11, the gene with the greatest expression change after PL treatment, for further study. Subsequently, RT-qPCR and Western blotting were employed to detect the expression of DDX11 in RCC cells treated with 2 and 5 µM of PL. The results indicated a significant decrease in both the mRNA and protein levels of DDX11 subsequent to PL treatment (Figure 3A,3B), which was consistent with the sequencing outcomes. We then detected the DDX11 expression in the RCC cell lines and normal renal epithelial cell line, HK-2. The result showed that HK-2 had the highest DDX11 expression, while 786-O and 769P cells had the lowest DDX11 expression (Figure 3C), suggesting that the expression level of DDX11 in RCC cells was negatively correlated with its sensitivity to PL. To clarify the role of DDX11 in PL-induced RCC cell death, we first verified the knockdown and overexpression efficiency of DDX11 (Figure 3D,3E). Next, we used CCK-8 and colony formation assays to investigate the role of DDX11 in PL’s inhibition of RCC cell proliferation. The results suggested that the overexpression of DDX11 significantly attenuated the inhibitory effect of PL on RCC cell proliferation (Figure 3F,3G). Therefore, we postulate that DDX11 is an important target of PL in inhibiting the progression of RCC.

DDX11 promoted miR-15b-3p maturation
Previous studies have revealed that DDX family proteins are involved in various biological processes, including alternative splicing, miRNA maturation, and mRNA nuclear export (10,17). In this study, we focused on the role of DDX11 in regulating miRNA maturation, which has not been reported on previously. We first conducted coimmunoprecipitation experiments and found that DDX11 can interact with the Drosha and DGCR8, which are critical to miRNA maturation (Figure 4A). To identify the miRNAs regulated by DDX11, we performed miRNA sequencing using a stable DDX11-overexpressed 786-O cell line and control cells (Figure 4B). Based on the miRNA sequencing results, we selected three miRNAs with the greatest change in expression level (miR-15b-3p, miR-562, and miR-466) for further experiments. Considering that the cleavage process from primary miRNA to precursor miRNA mainly depends on DGCR8 recognition and the cleavage action of Drosha (18), we first examined the effect of DDX11 overexpression on the expression of primary miRNA. As expected, overexpression of DDX11 did not affect the expression of primary miR-15b but did increase the expression of the miR-15b-3p precursor and mature forms (Figure 4C). Similarly, silencing of DDX11 did not affect the expression of primary miR-15b but did reduce the expression of the miR-15b-3p precursor and mature forms (Figure 4D). Furthermore, to verify that DDX11 promotes miR-15b-3p maturation via directly interacting with the Drosha and DGCR8, RIP–qPCR assays were performed. The results showed that DDX11 could only bind to primary miR-15b-3p but could not interact with the precursor or mature forms of miR-15b-3p (Figure 4E). Additionally, the DGCR8-RIP-qPCR assay indicated that silencing of DDX11 decreased the binding capacity between DGCR8 and primary miR-15b-3p, while DDX11 overexpression enhanced this binding capacity (Figure 4F). These findings confirmed that DDX11 promoted miR-15b-3p maturation via directly interacting with Drosha and DGCR8. Subsequently, DDX11-RIP-qPCR assays revealed that PL treatment impaired the interaction between DDX11 and primary miR-15b-3p (Figure 4G), which was consistent with our previous data. Moreover, qRT-PCR experiments confirmed a significant decrease in the expression of miR-15b-3p after PL treatment, and the overexpression of DDX11 was able to rescue the expression of miR-15b-3p (Figure 4H), indicating that PL regulates the expression of miR-15b-3p in RCC cells through DDX11.

PL inhibited RCC progression via regulating DDX11-miR-15b-3p
We then examined the role of miR-15b-3p in the proliferation of RCC cell under PL treatment. CCK-8 and colony formation assays demonstrated that the overexpression of DLD significantly alleviated the inhibitory effect of PL on the proliferation of RCC cells, while miR-15b-3p inhibition counteracted this effect (Figure 5A,5B). Furthermore, we measured the levels of ROS, MMP, and mitochondrial Fe2+ levels in miR-15b-3p-overexpressed cells under 2 µM of PL treatment. The results indicated that miR-15b-3p overexpression could counteract the effect of PL on the levels of ROS, Fe2+, and MMP (Figure 5C-5E).

DLD was regulated by the PL-DDX11-miR-15b-3p axis
In order to identify the downstream genes regulated by miR-15b-3p, we initially used the TargetScan website to predict potential miR-15b-3p target genes. Subsequently, by intersecting the predicted miRNA targets with our RNA-seq data, we selected three genes, namely PARPBP, PRB1, and DLD (Figure 6A). Considering that oxidative phosphorylation pathway ranked first in the KEGG pathway analysis of the transcriptome sequencing (Figure 2B) and that DLD figures prominently in energy metabolism (19), we initially considered DLD as a potential downstream gene of miR-15b-3p. Subsequently, through qRT-PCR and Western blot experiments, we found that downregulating miR-15b-3p increased the expression of DLD, while overexpressing miR-15b-3p inhibited the expression of DLD (Figure 6B,6C). Next, we used CCK-8 and colony formation assays to investigate the role of DLD in the proliferation of RCC cells under PL treatment. The results suggested that the overexpression of miR-15b-3p significantly mitigated the inhibitory effect of PL on the proliferation of RCC cells, while DLD overexpression counteracted this effect (Figure 6D-6F). Subsequently, we measured the levels of ROS, MMP, and mitochondrial Fe2+ levels in DLD-silenced cells under 2 µM of PL treatment. The results indicated that DLD knockdown could rescue the effect of PL on the levels of ROS, Fe2+, and MMP (Figure 6G-6I). These data suggest that DLD is regulated by the PL-DDX11-miR-15b-3p axis.

DDX11 as critical factor to the PL-induced inhibition of RCC
To determine the role of DDX11 in the PL-induced inhibition of RCC in vivo, a xenograft mice model was applied. Cells with stable overexpression of DDX11 or vector 769-P were subcutaneously injected into the BALB/c nude mice. The mice were then injected with PL (5 mg/kg) or vehicle intraperitoneally three times each week. After 6 weeks, the tumors were harvested, and the results showed that overexpression of DDX11 could significantly promote RCC proliferation under PL treatment (Figure 7A-7C). In addition, the IHC staining assays indicated that in the 769-P xenografts, the expression of DDX11 was negatively correlated with DLD (Figure 7D). These data demonstrate that DDX11 is the critical factor involved in the PL-induced inhibition of RCC.

Discussion
PL is a biologically active alkaloid extracted from natural product. Numerous studies have confirmed the pharmacological capabilities of PL as an anticancer agent (20). For instance, PL can limit the progression of breast cancer, colon cancer, oral cancer, thyroid cancer, and melanoma progression (21-24). However, the role of PL in RCC progression has not been extensively examined. In our study, we found PL inhibited the progression of RCC in a dose-dependent manner, supporting the value of PL in RCC treatment.
Previous studies have indicated that PL can increase the cellular ROS level through various mechanisms. Moreover, the excessive accumulation of ROS in cancer cells impairs the redox balance, further inducing oxidative stress and resulting in cancer cell death. In our study, we performed RNA-seq in 786-O cells with or without PL treatment. KEGG pathway analysis demonstrated that PL significantly regulated the oxidative phosphorylation and ferroptosis pathways. Moreover, PL significantly increased the levels of ROS and Fe2+ within the mitochondria, while reducing that of MMP, corroborating our RNA-seq data. Furthermore, we found that the morphology of mitochondria was impaired after treatment with PL. These data suggest that PL inhibits RCC proliferation by impairing mitochondrial homeostasis.
The DDX family has been widely reported to be involved various biological processes that are critical to tumor progression. However, the role of DDX11 in tumor progression has not been clarified in detail. Su et al. reported that DDX11 interacts with EZH2 to exert a protective effect against ubiquitination-mediated protein degradation (25). In our study, we found that PL treatment decreased DDX11 expression. Overexpression of DDX11 in PL-treated cells could rescue the phenotype of RCC cells both in vivo and in vitro. Furthermore, we found that DDX11 could promote the maturation of miR-15b-3p via binding to the key miRNA maturation proteins DGCR8 and Drosha. Additionally, functional assays showed that miR-15b-3p is involved in the PL/DDX5-induced RCC inhibition pathway. Meanwhile, t miR-15b-3p could regulate the expression of DLD, a metabolism-related gene which regulates redox balance in cells. Functional assay showed that PL impaired mitochondrial homeostasis by regulating the DDX11-miR-15b-3p-DLD axis.
Our study involved certain limitations which should be addressed. (I) In our study, we observed the regulation of DDX11 expression by PL both at the mRNA and protein levels. However, the details regarding the mechanism of this regulation were not examined. Subsequently, our forthcoming research will seek to clarify how exactly PL regulates DDX11 expression. (II) In our investigation of how DDX11 regulates DGCR8, we did not design corresponding truncations of DDX11 and DGCR8 to verify the specific binding domains responsible for their interaction. This step is crucial for determining the impact of their interaction on the maturation process of pre-miRNA. (III) We aim to further understand how DLD influences the mitochondrial homeostasis of RCC cells. While the regulation of ferroptosis by DLD has been documented in head and neck cancer, its specific role in RCC remains underexplored. Our subsequent work will focus on delineating the detailed regulatory mechanism of DLD in ferroptosis.
Conclusions
Our findings confirmed that PL significantly inhibits RCC progression through interfering with the mitochondrial homeostasis of RCC cells. Mechanistic analyses indicated that PL decreases the expression of DDX11, inhibits the maturation of miR-15b-3p, and further increases the level of DLD, resulting in a redox imbalance. Moreover, in vivo assays demonstrated that DDX11 plays a critical role in PL-induced RCC inhibition. In summary, our study indicates that DDX11 plays a critical role in the PL-induced inhibition of RCC by modulating the miR-15b-3p-DLD axis (Figure 8).

Acknowledgments
None.
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at https://tau.amegroups.com/article/view/10.21037/tau-2025-11/rc
Data Sharing Statement: Available at https://tau.amegroups.com/article/view/10.21037/tau-2025-11/dss
Peer Review File: Available at https://tau.amegroups.com/article/view/10.21037/tau-2025-11/prf
Funding: The study was supported by
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tau.amegroups.com/article/view/10.21037/tau-2025-11/coif). The authors have no conflicts of interest to declare.
Ethical Statement:
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Fuchs HE, et al. Cancer Statistics, 2021. CA Cancer J Clin 2021;71:7-33. [Crossref] [PubMed]
- Chen YW, Panian J, Rose B, et al. Recent Developments in the Management of Renal Cell Cancer. JCO Oncol Pract 2025; Epub ahead of print. [Crossref] [PubMed]
- Pham DX, Hsu T. Tumor-initiating and metastasis-initiating cells of clear-cell renal cell carcinoma. J Biomed Sci 2025;32:17. [Crossref] [PubMed]
- Huang RS, Chow R, Benour A, et al. Comparative efficacy and safety of ablative therapies in the management of primary localised renal cell carcinoma: a systematic review and meta-analysis. Lancet Oncol 2025;26:387-98. [Crossref] [PubMed]
- Giri VK, Zaemes J. The selection of targeted therapies for relapsed or refractory advanced renal cell carcinoma. Expert Rev Anticancer Ther 2025;25:337-49. [Crossref] [PubMed]
- Wali AF, Pillai JR, Talath S, et al. Phytochemicals in Breast Cancer Prevention and Treatment: A Comprehensive Review. Curr Issues Mol Biol 2025;47:30. [Crossref] [PubMed]
- Zhou J, Shu QJ, Wang T, et al. Piperlongumine induces ROS accumulation to reverse resistance of 5-FU in human colorectal cancer via targeting TrxR. Eur J Pharmacol 2025; Epub ahead of print. [Crossref] [PubMed]
- Li H, Wang M, Hua Z, et al. Piperlongumine alleviates viral myocarditis by inhibiting pyroptosis through NF-κB pathway. Phytomedicine 2025;140:156606. [Crossref] [PubMed]
- Winnard PT Jr, Vesuna F, Raman V. DExD-box RNA helicases in human viral infections: Pro- and anti-viral functions. Antiviral Res 2025;235:106098. [Crossref] [PubMed]
- Ma X, Lu T, Yang Y, et al. DEAD-box helicase family proteins: emerging targets in digestive system cancers and advances in targeted drug development. J Transl Med 2024;22:1120. [Crossref] [PubMed]
- Shen L, Zhang J, Xu M, et al. DDX3 acts as a tumor suppressor in colorectal cancer as loss of DDX3 in advanced cancer promotes tumor progression by activating the MAPK pathway. Int J Biol Sci 2022;18:3918-33. [Crossref] [PubMed]
- Yang Y, Yu S, Liu W, et al. Ferroptosis-related signaling pathways in cancer drug resistance. Cancer Drug Resist 2025;8:1. [Crossref] [PubMed]
- Xu W, Guan G, Yue R, et al. Chemical Design of Magnetic Nanomaterials for Imaging and Ferroptosis-Based Cancer Therapy. Chem Rev 2025;125:1897-961. [Crossref] [PubMed]
- Yang G, Qian B, He L, et al. Application prospects of ferroptosis in colorectal cancer. Cancer Cell Int 2025;25:59. [Crossref] [PubMed]
- Li L, Xu Y, Yang W, et al. Construction of a two-gene prognostic model related to ferroptosis in renal cell carcinoma. Transl Androl Urol 2023;12:1167-83. [Crossref] [PubMed]
- Chang K, Chen Y, Zhang X, et al. DPP9 Stabilizes NRF2 to Suppress Ferroptosis and Induce Sorafenib Resistance in Clear Cell Renal Cell Carcinoma. Cancer Res 2023;83:3940-55. [Crossref] [PubMed]
- Li S, Feng T, Yuan H, et al. DEAD-box RNA helicases in the multistep process of tumor metastasis. Mol Biol Rep 2024;51:1006. [Crossref] [PubMed]
- Kim H, Lee YY, Kim VN. The biogenesis and regulation of animal microRNAs. Nat Rev Mol Cell Biol 2025;26:276-96. [Crossref] [PubMed]
- Yan LJ, Wang Y. Roles of Dihydrolipoamide Dehydrogenase in Health and Disease. Antioxid Redox Signal 2023;39:794-806. [Crossref] [PubMed]
- Modi SR, Andey T. Piperlongumine in combination with EGFR tyrosine kinase inhibitors for the treatment of lung cancer cells. Oncol Res 2024;32:1709-21. [Crossref] [PubMed]
- Jamali F, Lan K, Daniel P, et al. Synergistic Dual Targeting of Thioredoxin and Glutathione Systems Irrespective of p53 in Glioblastoma Stem Cells. Antioxidants (Basel) 2024;13:1201. [Crossref] [PubMed]
- Dai Y, Chen J, Fang J, et al. Piperlongumine, a natural alkaloid from Piper longum L. ameliorates metabolic-associated fatty liver disease by antagonizing the thromboxane A(2) receptor. Biochem Pharmacol 2024;229:116518. [Crossref] [PubMed]
- Zhang M, Wang K, Li M, et al. Highly Efficient and Long-Lasting Chemiluminescence-Functionalized Nanohydrogel for Imaging-Guided Precise Piperlongumine Chemotherapy. Anal Chem 2024;96:19833-9. [Crossref] [PubMed]
- Qiu J, Guo F, Shi J, et al. Piperlongumine inhibits glioblastoma proliferation by inducing ferroptosis. J Pharm Pharmacol 2024;rgae148. [Crossref] [PubMed]
- Su SG, Li QL, Zhang MF, et al. An E2F1/DDX11/EZH2 Positive Feedback Loop Promotes Cell Proliferation in Hepatocellular Carcinoma. Front Oncol 2020;10:593293. [Crossref] [PubMed]
(English Language Editor: J. Gray)


