Nitrocellulose tissue prints: an innovative approach to preparing high quality DNA and RNA from prostate biopsies without compromising the cores for pathology diagnosis
Biopsy-based prostate biomarker research: opportunities and challenges
Much of the research that has been done on prostate cancer tissue biomarkers has relied on radical prostatectomies for biospecimens. Although this is a logical choice for studies of organ confined prostate cancer, the fact that most prostate cancers are not visibly evident in fresh surgical specimens can make collection of fresh-frozen tumor tissues somewhat challenging. Moreover, it is well recognized that in biorepository radical prostatectomy specimen collections, important groups of patients are under-represented or missing entirely. Men who are diagnosed with high volume and/or high-grade prostate cancer are often treated with hormonal and radiation therapy rather than surgery; for these high-risk patients there are no radical prostatectomy specimens. In addition, at many centers, including the University of Alabama at Birmingham, a preference of African American patients for non-surgical treatment has been a factor in creating an unintentional under-representation of this important patient population in radical prostatectomy collections. One of the major priorities in prostate cancer biomarker research is to ensure that high-risk patients and African American patients are well represented in the datasets that inform our thinking about prostate cancer biology.
Using prostate biopsy tissues for molecular biomarker research significantly expands the range of available patients compared to studies that rely only on radical prostatectomy specimens (1,2). These include men whose biopsies show no cancer and men diagnosed with lower risk disease who choose active surveillance as well as men who are treated non-surgically. However, one of the challenges of biopsy-based biomarker research is the limited amount of tissue that can be obtained from each core. A prostate biopsy core is approximately the size of half a toothpick. Clinical care must come first, and the priority for prostate biopsy specimens is a pathologic diagnosis. Frequently, after biopsy cores have been fixed in formalin, embedded in paraffin and sectioned at several levels to produce slides for histopathology, only a narrow sliver of residual formalin-fixed paraffin-embedded (FFPE) tissue remains in the block as a potential research specimen. The introduction of clinical biomarker tests that use prostate cancer tissue is creating new challenges. At many centers, the demand for additional sections for clinical molecular testing has increased to the point where access to residual FFPE biopsy tissue for research must be restricted or prohibited.
Tissue print technologies as a practical approach to prostate biopsy biomarker research
To meet the challenges of biopsy-based tissue biomarker research, we have developed and fully implemented innovative tissue print technologies that allow us to obtain high quality RNA and DNA from each biopsy core without compromising the tissue specimen for pathology processing and diagnosis. Tissue prints are nitrocellulose touch preps that combine a classic biological research technique with contemporary methods for molecular biomarker analysis. With larger specimens such as radical prostatectomies, tissue prints provide a practical approach to characterizing molecular biomarkers at surgical margins (3) and mapping tissue slices for pathology or imaging correlations (4). With prostate biopsies, tissue prints support molecular biomarker studies of valuable specimens that may otherwise be significantly limited or entirely unavailable for research (5,6).
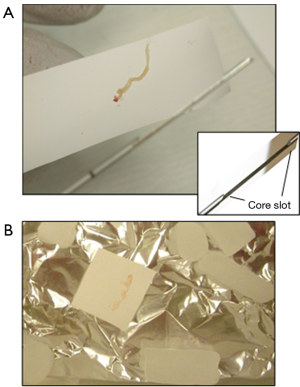
The collection of prostate biopsy tissue prints is illustrated in Figures 1,2. Prostate biopsy is usually an outpatient procedure and we have incorporated biopsy tissue print collection into the routine clinical workflow of these outpatient-based biopsy sessions. Because only one needle is used for a biopsy procedure and 12 or more cores may be obtained, each core must be transferred in turn from the cutting needle to a jar of formalin fixative. A tissue print is collected by using a nitrocellulose paper to transfer the tissue from the cutting needle to the fixation jar. Of importance, blotting the core with nitrocellulose paper does not dry or cause other artifacts in the biopsy tissue. The tissue core is then immersed in formalin fixative as usual and submitted for pathology diagnosis; the tissue print is snap frozen and sent to the laboratory where it will be processed for biomarker studies. The imprint of the biopsy core is visible on the nitrocellulose paper, confirming the success of collection (Figure 2). In some cases, during the transfer, one end of the print and core are marked with ink to facilitate orientation of the tissue print with the H&E sections produced from the core during pathology processing. This allows the processing lab to trim the tissue print to enrich for areas of cancer or other regions of interest.
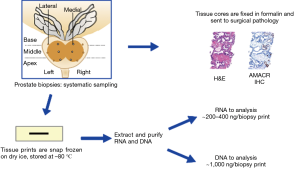
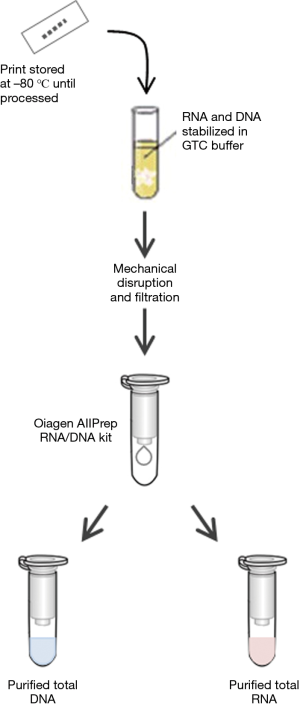
We routinely prepare purified RNA and DNA from each biopsy tissue print using a commercially available kit (Qiagen AllPrep DNA/RNA) with a modified protocol (Figure 3). The approach to processing tissue prints for RNA and DNA biomarkers is based on techniques used to purify RNA and DNA from fibrous tissue. Cellular material is solubilized into a nucleic acid stabilizing buffer using a bead mill such as a Retsch Qiagen TissueLyser or similar unit. The nitrocellulose fibers are then removed by filtration before beginning RNA and DNA purification. The yield of purified RNA and DNA from each tissue print depends in part on the cellular composition of the tissue in the biopsy core. For tissue prints from biopsy cores that contain >50% high-grade prostate cancer, the median yields of purified RNA and DNA are approximately 480 and 940 ng, respectively. For tissue prints of cores with no cancer, the median RNA and DNA yields are approximately 250 and 920 ng, respectively. Global gene expression analysis of biopsy “print-core” sample pairs using Affymetrix arrays shows excellent gene expression concordance between a tissue print and its corresponding biopsy core (Pearson 0.96–0.98; R2 approximately 0.92–0.96). Because the tissue prints are snap frozen rather than fixed in formalin, the purified RNA and DNA is of high quality and suitable for advanced biomarker analysis; prostate biopsy tissue print samples have been successfully utilized for mRNA and microRNA gene expression profiling, genotyping, DNA methylation and sequencing analyses (8-12). Proteins can also be extracted from biopsy tissue prints either by using a commercial kit (Qiagen AllPrep DNA/RNA/Protein, for example) or by electrophoresis (see “print-phoresis” in reference 3). Print-phoresis is particularly well suited for protein immunoblot analysis (“western blots”) and for preparation of samples for protein mass spectroscopy.
Expanding opportunities for biopsy-based prostate biomarker studies
To date we have collected and processed biopsy tissue prints from more than 750 prostate biopsy procedures, including standard-of-care systematic biopsies and MRI-guided biopsy procedures. While correlations with pathology are a key component of each biomarker study, molecular annotations of MRI features are an increasingly important objective. In addition, we have begun to take advantage of the fact that the cells that are transferred from fresh prostate biopsy and surgical specimens to nitrocellulose are viable and can be used for live cell studies. As platforms for analyzing viable single cells become increasingly sophisticated, the opportunities to re-format tissue prints to study tumor metabolism and drug response are expanding. In summary, tissue prints provide a practical approach to biopsy-based prostate cancer studies that is fully implemented for molecular biomarker research and well positioned for emerging technologies that can characterize viable cells.
Acknowledgements
Funding: This work was supported by research grants from the NIH and DOD to Dr. Sandra M. Gaston (21 CA112220 and W81XWH-10-1-0544) and to Dr. William E. Grizzle (UM1CA183728 and W8XWH-10-1-0543).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Gaston SM, Otali D, Kearns J, et al. A biopsy-focused approach to molecular studies of prostate cancer in African Americans: the Birmingham Alabama Prostate Cancer Consortium (BAPrCa). Ninth AACR Conference on the Science of Cancer Health Disparities in Racial/Ethnic Minorities and the Medically Underserved; September 2016.
- Gaston SM, Kolettis PN, Bryant JE, et al. Implementation of prostate biopsy tissue print technologies for molecular biomarker studies. Annual Meeting of the American Urological Association; May 2017.
- Gaston SM, Soares MA, Siddiqui MM, et al. Tissue-print and print-phoresis as platform technologies for the molecular analysis of human surgical specimens: mapping tumor invasion of the prostate capsule. Nat Med 2005;11:95-101. [Crossref] [PubMed]
- Lenkinski RE, Bloch BN, Liu F, et al. An illustration of the potential for mapping MRI/MRS parameters with genetic over-expression profiles in human prostate cancer. MAGMA 2008;21:411-21. [Crossref] [PubMed]
- Gaston SM, Kearns J, Adams GW, et al. Satisfying the fatty acid demand of prostate cancer: Outlier overexpression of genes involved in de novo synthesis vs fatty acid uptake defıne two different and potentially synergistic prostate cancer phenotypes. Annual Meeting of the American Association for Cancer Research; April 2017.
- Van Neste L, Groskopf J, Grizzle WE, et al. Epigenetic risk score improves prostate cancer risk assessment. Prostate 2017;77:1259-64. [Crossref] [PubMed]
- Westring CG, Kristinsson R, Gilbert DM, et al. Validation of reduced-scale reactions for the Quantifiler Human DNA kit. J Forensic Sci 2007;52:1035-43. [Crossref] [PubMed]
- Gaston SM, Vu D, Brice MJ, et al. Tissue Print Profiling of Prostate Needle Biopsies: Obtaining Comprehensive Tissue Sampling for Molecular Marker Analysis without Compromising Microscopic Evaluation of the Biopsy Cores. J Urol 2004;171:483. [Crossref]
- Wang F, Wang L, Briggs C, et al. DNA degradation test predicts success in whole-genome amplification from diverse clinical samples. J Mol Diagn 2007;9:441-51. [Crossref] [PubMed]
- Gaston SM, Guerra AL, Nadadur R, et al. Overexpression of miR-17 family miRNAs in prostate cancer biopsies: Evidence for a stem-cell-like miRNA profile in high grade/high stage tumors. Cancer Research 2010;70:3049. [Crossref]
- Angelucci A, Pace G, Sanità P, et al. Tissue print of prostate biopsy: a novel tool in the diagnostic procedure of prostate cancer. Diagn Pathol 2011;6:34. [Crossref] [PubMed]
- Gaston SM, Grizzle WE, Kittles RA, et al. The use of innovative prostate biopsy tissue print techniques for molecular genomic, epigenomic and gene expression studies. Cancer Research 2015;75:SY29-02. [Crossref]


