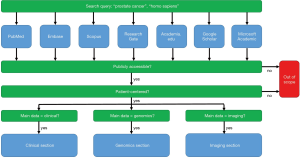
An overview of publicly available patient-centered prostate cancer datasets
Introduction
Prostate cancer (PCa) is the second most common cancer in men, and the second leading cause of death from cancer in men (1). Many studies on PCa have been carried out, each taking much time before the data is collected and ready to be analyzed. The datasets created in these studies are usually collected by academic institutes, who are often unwilling to share the data because of concerns over ownership, publications or patient consent. Because of the new privacy regulations in the EU General Data Protection Regulation (GDPR), this data sharing is becoming increasingly more difficult (2). However, on the internet a wide range of PCa datasets are already available, ready to be used for data mining and analysis. Some of them are well-known to researchers in the field, but others remain hidden because they were published in a low-impact journal or are simply not on the first page of Google. Nevertheless, these datasets could be still used for data mining, predictive modelling or other purposes, reducing the need to setup new studies to collect data. In the current scientific climate, moving more and more to the analysis of ‘big data’ (3,4) and large, international, multi-site projects using a modern IT infrastructure, such as Movember GAP3 (5), ERSPC (6) and PCMM (7), these datasets could be proven extremely valuable.
This review presents an overview of publicly available patient-centered PCa datasets (8), divided into three categories (clinical, genomics and imaging) and an ‘overall’ section to enable researchers to select a suitable dataset for analysis, without having to go through days of work to find the right data.
The ‘Clinical data’ section contains datasets that have a number of clinical parameters, i.e., data that can be captured in numerical or text fields. In the area of PCa these are, for example: age, Gleason scores, TNM stages and PSA values, but also values derived from the genomics and imaging domains, such as biomarker expression values and PI-RADS scores (9).
The ‘Genomics data’ section describes a number of datasets resulting from genomics studies, such as microarray experiments. Websites like cBioPortal (10), GEO (11) and ArrayExpress (12) and apps like camcAPP (13) can be used to browse through genomics datasets.
In the ‘Imaging data’ section, a number of data sources containing Magnetic Resonance (MR), UltraSound (US), Positron-emission tomography (PET), Computed Tomography (CT) and histopathology images are listed.
The ‘Overall’ section brings all datasets within this review together, and shows which datasets contain information from more than one domain (clinical/genomics/imaging). It gives a complete picture of all publicly available patient-centered PCa datasets.
Methods
Scientific literature databases and academic social network sites such as PubMed/Medline, Embase, Scopus, ResearchGate, Academia.edu, Google Scholar and Microsoft Academic were searched to acquire a list of human PCa databases (Figure 1). We also used the information from other reviews (14) in this paper. All databases in the combined list were then checked for public availability. Only databases that were either directly publicly available or available after signing a research data agreement or retrieving a free login were selected for inclusion in this review. Data should be available to commercial parties as well. This paper focuses on patient-centered data, so the genomics data section does not include gene-centered databases, pathway-centered databases, etc. that are not linked to patients. This exclusion ensures that the genomics data can be more easily combined with the clinical and imaging data.
Results
Clinical data
The Prostate, Lung, Colorectal and Ovarian (PLCO) Cancer Screening Trial (15) is a randomized, controlled trial to determine whether certain screening exams reduce mortality from prostate, lung, colorectal and ovarian cancer. A total of 76,682 male participants were enrolled between November 1993 and July 2001. Data collected up to December 31, 2009 for the first 13 years of participation for each subject in the PLCO trial are available at https://biometry.nci.nih.gov/cdas/datasets/plco/20/. Six PCa screening datasets are available:
- The Prostate dataset is a comprehensive dataset that contains nearly all the PLCO study data available for PCa screening, incidence, and mortality analyses. The dataset contains one record for each of the 76,682 male participants in the PLCO trial.
- The Prostate Screening dataset (177,315 records, 35,875 subjects, one record per year of screening) contains additional information from PSA and Digital Rectal Exam (DRE) cancer screens. This includes details of the blood draw, QA DRE results, reasons for inadequate exams, and additional findings that were not suspicious for cancer.
- The Prostate Screening Abnormalities dataset (10,527 records, 5,743 subjects, one record per abnormality) contains information for each induration found during the DRE screen. This includes the location, type, size, grade, and extent of each induration.
- The Prostate Diagnostic Procedures dataset (95,837 records, 15,307 subjects, one record per procedure) contains information about the diagnostic procedures prompted by positive PCa screens, as well as diagnostic/staging procedures associated with any PCa diagnosed during the 13 years of follow-up.
- The Prostate Medical Complications dataset (3,350 records, 2,164 subjects, one record per medical complication) contains information about the medical complications caused by diagnostic workup for PCa.
- The Prostate Treatments dataset (13,409 records, 7,614 subjects, one record per treatment procedure) contains specifics of the initial treatment following the diagnosis of PCa.
The Surveillance, Epidemiology, and End Results (SEER) database (16) of the National Cancer Institute at https://seer.cancer.gov/data/seerstat/nov2017/ provides information on cancer statistics in an effort to reduce the cancer burden among the U.S. population. SEER is supported by the Surveillance Research Program, which provides national leadership in the science of cancer surveillance as well as analytical tools and methodological expertise in collecting, analyzing, interpreting, and disseminating reliable population-based statistics. The SEER research data include SEER incidence and population data associated by age, sex, race, year of diagnosis, and geographic areas (including SEER registry and county). SEER research data are released every Spring based on the previous November’s submission of data. A research data agreement needs to be signed and approved before the data can be accessed. The SEER PCa dataset consist of four parts:
- YR1973_2015.SEER9 contains the SEER November 2017 Research Data files from nine SEER registries (Atlanta, Connecticut, Detroit, Hawaii, Iowa, New Mexico, San Francisco-Oakland, Seattle-Puget Sound, and Utah) for 1973-2015 (n=637005).
- YR1992_2015.SJ_LA_RG_AK contains the SEER November 2017 Research Data files from the San Jose-Monterey, Los Angeles, Rural Georgia and Alaska Natives SEER registries for 1992-2015 (n=164576).
- YR2000_2015.CA_KY_LO_NJ_GA contains the SEER November 2017 Research Data files from the Greater California, Kentucky, Louisiana, New Jersey, and Greater Georgia SEER registries for 2000-2015 (n=461,552).
- YR2005.LO_2ND_HALF contains the July—December 2005 diagnoses for Louisiana from their November 2017 SEER submission (n=1,352).
The National Program of Cancer Registries (NPCR) offers access to two public use databases at https://www.cdc.gov/cancer/npcr/public-use/ in collaboration with SEER. The databases include data by demographic characteristics (for example, age, sex, race, and year of diagnosis) and tumor characteristics (for example, site, histology, stage, and behavior). Hospitals, physicians, and laboratories across the nation report these data to central cancer registries supported by CDC and NCI. The two databases are:
- 2001–2015 database (17), which includes data for 50 states and the District of Columbia (n=3,086,534 for PCa).
- 2005–2015 database (18), which includes data for 50 states, the District of Columbia, and Puerto Rico (n=2,294,444 for PCa).
The popular statistical package R contains a PCa dataset from Stamey et al. [1989] (19), available for analysis when using the ElemStatLearn package. It contains data from 97 patients for 9 clinical variables. More information can be found at https://cran.r-project.org/web/packages/ElemStatLearn/ElemStatLearn.pdf.
Genomics data
The popular tool cBioPortal (10), a web portal for cancer genomics data, offers access to sixteen PCa datasets (including clinical and biospecimen data in some cases). cBioPortal has several built-in visualizations and analyses of the genomics data, which make it very easy to explore the data without much effort. The datasets, available at http://www.cbioportal.org/datasets, are:
- Genomic Hallmarks of Prostate Adenocarcinoma (CPC-GENE) (20). Comprehensive genomic profiling of 477 Prostate Adenocarcinoma samples from CPC-GENE and public data sets, including TCGA-PRAD. Data available at http://www.cbioportal.org/study?id=prad_cpcg_2017.
- MSK-IMPACT Clinical Sequencing Cohort (MSKCC): PCa (21). Targeted sequencing of clinical cases via MSK-IMPACT for PCa. Data available at http://www.cbioportal.org/study?id=prad_mskcc_2017.
- Metastatic Prostate Adenocarcinoma (MCTP) (22). Comprehensive profiling of 61 PCa samples, including 50 metastatic CRPCs and 11 high-grade localized PCa. Generated by Arul Chinnaiyan's and Scott Tomlins' labs at the University of Michigan. Data available at http://www.cbioportal.org/study?id=prad_mich.
- Metastatic Prostate Cancer, SU2C/PCF Dream Team (23). Comprehensive analysis of 150 metastatic PCa samples by the SU2C/PCF Dream Team. Data available at http://www.cbioportal.org/study?id=prad_su2c_2015.
- Neuroendocrine Prostate Cancer (Trento/Cornell/Broad) (24). Whole exome and RNA Seq data of castration resistant adenocarcinoma and castration resistant neuroendocrine PCa (somatic mutations and copy number aberrations, 114 samples). Data available at http://www.cbioportal.org/study?id=nepc_wcm_2016.
- Prostate Adenocarcinoma (Broad/Cornell 2013) (25). Comprehensive profiling of 57 PCa samples. Generated by Levi Garraway’s lab at the Broad Institute and Mark Rubin’s lab at Cornell. Data available at http://www.cbioportal.org/study?id=prad_broad_2013.
- Prostate Adenocarcinoma (Broad/Cornell 2012) (26). Comprehensive profiling of 112 PCa samples. Generated by Levi Garraway’s lab at the Broad Institute and Mark Rubin’s lab at Cornell. Data available at http://www.cbioportal.org/study?id=prad_broad.
- Prostate Adenocarcinoma (Sun Lab) (27). Whole-genome and Transcriptome Sequencing of 65 Prostate Adenocarcinoma Patients. Generated by the Sun Lab 2017. Data available at http://www.cbioportal.org/study?id=prad_eururol_2017.
- Prostate Adenocarcinoma (Fred Hutchinson CRC) (28). Comprehensive profiling of PCa samples. Generated by Peter Nelson's lab at the Fred Hutchinson Cancer Research Center. Data available at http://www.cbioportal.org/study?id=prad_fhcrc.
- Prostate Adenocarcinoma (MSKCC) (29). MSKCC Prostate Oncogenome Project. 181 primary, 37 metastatic PCa samples, 12 PCa cell lines and xenografts. Data available at http://www.cbioportal.org/study?id=prad_mskcc.
- Prostate Adenocarcinoma (MSKCC/DFCI) (30). Whole Exome Sequencing of 1013 PCa samples. Data available at http://www.cbioportal.org/study?id=prad_p1000.
- Prostate Adenocarcinoma (TCGA) (31). Integrated profiling of 333 primary prostate adenocarcinoma samples. Data available at http://www.cbioportal.org/study?id=prad_tcga_pub.
- Prostate Adenocarcinoma (TCGA, PanCancer Atlas) (32). Comprehensive TCGA PanCanAtlas data from 11k cases and all TCGA tumor types (33). Data available at http://www.cbioportal.org/study?id=prad_tcga_pan_can_atlas_2018.
- Prostate Adenocarcinoma (TCGA, Provisional). TCGA Prostate Adenocarcinoma (499 samples). Data available at http://www.cbioportal.org/study?id=prad_tcga.
- Prostate Adenocarcinoma CNA study (MSKCC) (33). Copy-number profiling of 103 primary PCa samples from MSKCC. Data available at http://www.cbioportal.org/study?id=prad_mskcc_2014.
- Prostate Adenocarcinoma Organoids (MSKCC) (34). Exome profiling of PCa samples and matched organoids (12 samples). Data available at http://www.cbioportal.org/study?id=prad_mskcc_cheny1_organoids_2014.
A subsite of cBioPortal, http://www.cbioportal.org/genie, contains data from the Genomics Evidence Neoplasia Information Exchange (GENIE) project (35) of the American Association for Cancer Research (AACR). The GENIE project seeks to identify and validate genomic biomarkers relevant to cancer treatment by linking tumor genomic data from clinical sequencing efforts with longitudinal clinical outcomes. The project includes data from eleven cancer centers from the USA (7×), Canada, the Netherlands, France and the United Kingdom. GENIE version 5.0 contains data from 2,214 PCa samples (from 2,008 patients): 2,172× Prostate Adenocarcinoma, 28× Prostate Neuroendocrine Carcinoma, 13× Prostate Small Cell Carcinoma and 1× Prostate Squamous Cell Carcinoma. The data is also accessible through https://www.synapse.org/genie.
The International Cancer Genome Consortium (ICGC) Data Portal (36) currently contains six PCa datasets, which can be found at https://dcc.icgc.org/q?q=prostate&type=project:
- PRAD-CA (37) (125 subjects). Prostate Adenocarcinoma—Canada. Collected by the CPC-GENE network and connected to the 1st dataset in cBioportal mentioned above. Data available at https://dcc.icgc.org/projects/PRAD-CA.
- PRAD-FR (38) (25 subjects). Prostate Adenocarcinoma—France. Collected by ten French and one Spanish research organization. Data available at https://dcc.icgc.org/projects/PRAD-FR.
- EOPC-DE (39) (211 subjects), Early Onset Prostate Cancer—Germany. Collected by six German research organizations. Data available at https://dcc.icgc.org/projects/EOPC-DE.
- PRAD-UK (40) (216 subjects). Prostate Adenocarcinoma—United Kingdom. Collected by the international Cancer Research UK funded Prostate Cancer Network (CR-UKPCN). Data available at https://dcc.icgc.org/projects/PRAD-UK.
- PRAD-US (41) (500 subjects). Prostate Adenocarcinoma TCGA—United States. Collected by sixteen American and one Canadian research organization. Data available at https://dcc.icgc.org/projects/PRAD-US.
- PRAD-CN (27,42) (65 subjects). Prostate Cancer—China. Collected by the Sun Lab. The same as the 8th dataset in cBioportal mentioned above. Data available at https://dcc.icgc.org/projects/PRAD-CN.
The Genomics Data Commons (GDC) (43) gives access to The Cancer Genome Atlas Prostate Adenocarcinoma (TCGA-PRAD) dataset, which is the same as the 14th dataset in cBioPortal mentioned above, and the 5th dataset in the ICGC Data Portal. It can be accessed at https://portal.gdc.cancer.gov/projects/TCGA-PRAD.
The Gene Expression Omnibus (GEO) database (11) from the National Center for Biotechnology Information (NCBI) contains 51 curated human PCa datasets, which can be retrieved at https://www.ncbi.nlm.nih.gov/gds/?term=%E2%80%9Cprostate+cancer%E2%80%9D%5BTitle%5D+AND+%22Homo+sapiens%22%5Bporgn%3A__txid9606%5D+AND+gds%5BFilter%5D (Table S1). These are only the curated datasets; there are in total 834 human PCa series available in GEO.
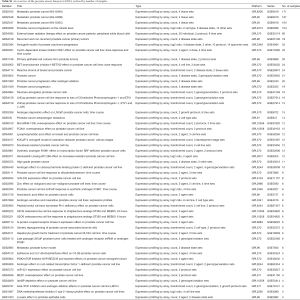
Full table
The ArrayExpress database (12) from the European Bioinformatics Institute (EBI) contains 126 human PCa datasets. We used the “ArrayExpress data only” checkbox to avoid datasets that are in GEO as well. The list of datasets can be retrieved at https://www.ebi.ac.uk/arrayexpress/browse.html?keywords=prostate+cancer&organism=Homo+sapiens&directsub=on (Table S2).
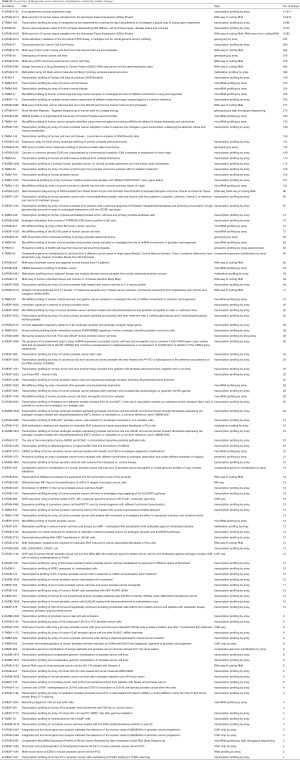
Full table
Imaging data
The Cancer Imaging Archive (TCIA) (44) has nine PCa datasets available, which can be found at http://www.cancerimagingarchive.net/:
- The Prostate-MRI collection (26 subjects) (45) of prostate Magnetic Resonance Images (MRIs) was obtained with an endorectal and phased array surface coil at 3T (Philips Achieva). Each patient had biopsy confirmation of cancer and underwent a robotic-assisted radical prostatectomy. A mold was generated from each MRI, and the prostatectomy specimen was first placed in the mold, then cut in the same plane as the MRI. The data was generated at the National Cancer Institute, Bethesda, Maryland, USA between 2008 and 2010, and can be downloaded from https://wiki.cancerimagingarchive.net/display/Public/PROSTATE-MRI (limited access).
- In the Prostate-Diagnosis project (92 subjects) (46), PCa T1- and T2-weighted magnetic resonance images (MRIs) were acquired on a 1.5 T Philips Achieva by combined surface and endorectal coil, including dynamic contrast-enhanced images obtained prior to, during and after I.V. administration of 0.1 mmol/kg body weight of Gadolinium-DTPA (pentetic acid). Data is available at https://wiki.cancerimagingarchive.net/display/Public/PROSTATE-DIAGNOSIS.
- NaF Prostate (9 subjects) (47,48) is a collection of F-18 NaF positron emission tomography/computed tomography (PET/CT) images in patients with PCa, with suspected or known bone involvement. This dataset is available for download at https://wiki.cancerimagingarchive.net/display/Public/NaF+Prostate.
- The Prostate-3T project (64 subjects) (49) provided imaging data to TCIA as part of an ISBI challenge competition in 2013. Prostate transversal T2-weighted magnetic resonance images (MRIs) acquired on a 3.0T Siemens TrioTim using only a pelvic phased-array coil were acquired for PCa detection. Data can be downloaded from https://wiki.cancerimagingarchive.net/display/Public/Prostate-3T.
- The QIN PROSTATE collection (22 subjects) (50,51) of the Quantitative Imaging Network (QIN) contains multiparametric MRI images collected for the purposes of detection and/or staging of PCa . The MRI parameters include T1- and T2-weighted sequences as well as Diffusion Weighted and Dynamic Contrast-Enhanced MRI. The images were obtained using endorectal and phased array surface coils at 3.0T (GE Signa HDx 15.0) The value of this collection is to provide clinical image data for the development and evaluation of quantitative methods for PCa characterization using multiparametric MRI. Data can be accessed, after a request, through https://wiki.cancerimagingarchive.net/display/Public/QIN+PROSTATE (limited access).
- The TCGA-PRAD project (14 subjects) (52), also mentioned in the Genomics section of this review, also has imaging data (CT, PT, MR and pathology images) available, which can be accessed through https://wiki.cancerimagingarchive.net/display/Public/TCGA-PRAD. It also contains a link to the clinical data belonging to this study.
- The Prostate Fused-MRI-Pathology collection (28 subjects) (53) is a combination of MRI images and histopathology slides. It comprises a set of 3 Tesla T1-weighted, T2-weighted, Diffusion weighted and Dynamic Contrast Enhanced prostate MRI along with accompanying digitized histopathology (H&E stained) images of corresponding radical prostatectomy specimens. The MRI scans also have a mapping of extent of PCa on them. The dataset is accessible at https://wiki.cancerimagingarchive.net/display/Public/Prostate+Fused-MRI-Pathology.
- The PROSTATEx Challenge dataset (346 subjects) (54,55) is a retrospective set of prostate MR studies. All studies included T2-weighted (T2W), proton density-weighted (PD-W), dynamic contrast enhanced (DCE), and diffusion-weighted (DW) imaging. Data can be downloaded at https://wiki.cancerimagingarchive.net/display/Public/SPIE-AAPM-NCI+PROSTATEx+Challenges.
- The QIN-PROSTATE-Repeatability dataset (15 subjects) (56-58) is a dataset with multiparametric prostate MRI applied in a test-retest setting, allowing to evaluate repeatability of the MRI-based measurements in the prostate. The imaging data is accompanied by two types of derived data: (I) manual segmentations of the total prostate gland, peripheral zone of the prostate gland, suspected tumor and normal regions (where applicable) and (II) volume measurements (for axial T2w images and ADC images) and mean ADC (for ADC images) corresponding to the segmented regions. Data can be accessed, after a request, through https://wiki.cancerimagingarchive.net/display/Public/QIN-PROSTATE-Repeatability.
Overall
The above sections show all clinical datasets, genomics datasets and imaging datasets. The most valuable datasets however are those that consist of a combination of these three domains, because it enables researchers to study connections, determine correlations, etc. Table 1 shows a combined overview of the clinical, genomics and imaging datasets. There is only one dataset that has data from all three domains: the TCGA dataset (52) [also known as PRAD-US (41)]. Furthermore, there are 20 clinical + genomics datasets and 1 clinical + imaging dataset. The full list of URLs from which each dataset can be downloaded, has been submitted to the Awesome Public Datasets list at https://github.com/awesomedata/awesome-public-datasets#prostatecancer.
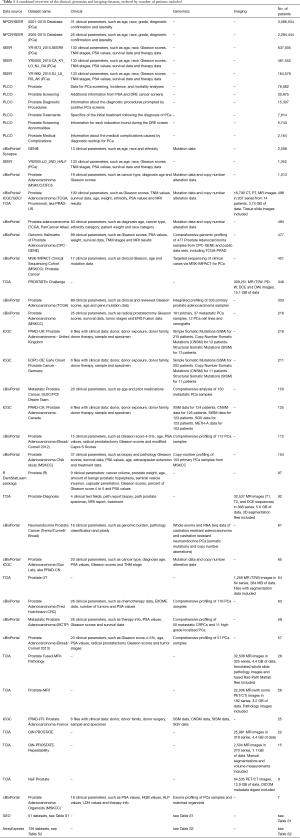
Full table
Discussion
Despite all the attention that has been given to making this overview of publicly available databases as extensive as possible, it is very likely not complete, and will also be outdated soon. However, this review might help many PCa researchers to find suitable datasets to answer the research question with, without the need to start a new data collection project. In the coming era of big data analysis and precision medicine (4), overviews like this are becoming more and more useful, and even necessary because of stricter privacy regulations (2). In the shift to data-driven research, the focus should be on data quality, as researchers depend more and more on the data not only for analysis, but also to generate hypotheses. The large amounts of data make it more difficult to do manual quality control, increasing the need for data quality control software. The datasets discussed within this overview seem to be of high quality, although it should be noted that some non-PCa-specific datasets such as the SEER and NPCR database, needed quite a lot of decoding work (i.e., translating codes to their PCa-specific description), increasing the risk of human errors. The SEER database, which started in 1973, also has some legacy issues (e.g., containing different versions of cancer staging scores). It should be noted as well that most datasets do not adhere to the FAIR (Findability, Accessibility, Interoperability, Reusability) guiding principles for scientific data management and stewardship (59), but this could be expected since almost all datasets were generated before these principles were published. Hopefully they will be FAIRified in the near future. Some datasets in this overview contain only a small number of patients, such as the NaF Prostate study and the Prostate Adenocarcinoma Organoids (MSKCC) study. In these cases it might be useful to combine datasets, to get to a higher sample size (and statistical power) by manual or automated data model mapping (60). It might also be useful for scientists to have access to the original biomaterial from which the data was derived. Therefore, the Prostate Cancer Biorepository Network (61) is an interesting initiative: its goal is to develop a biorepository with high quality, well-annotated specimens obtained in a systematic, reproducible fashion using optimized and standardized protocols. It is a collaboration between six U.S. academic institutes and the U.S. department of defense. Finally, the success of big data analysis does not only depend on access to data and/or biospecimens, but also on the collaboration between field experts (urologists, but also imaging and genomics experts) and IT experts (62). There are very little people that have an in-depth knowledge about the disease area, the used techniques, data integration and data analysis, which is why multi-disciplinary research teams are a must in this ‘big data’ age.
Acknowledgements
The author thanks Chris Bangma for presenting a poster version of this manuscript at the AUA 2018 (8). The NPCR/SEER data were provided by central cancer registries participating in CDC’s National Program of Cancer Registries (NPCR) and/or NCI’s Surveillance, Epidemiology, and End Results (SEER) Program and submitted to CDC and NCI in November, 2017. The author thanks the National Cancer Institute for access to NCI’s data collected by the Prostate, Lung, Colorectal and Ovarian (PLCO) Cancer Screening Trial. The statements contained herein are solely those of the author and do not represent or imply concurrence or endorsement by NCI. The author would like to acknowledge the American Association for Cancer Research and its financial and material support in the development of the AACR Project GENIE registry, as well as members of the consortium for their commitment to data sharing. Interpretations are the responsibility of study author. The results on the TCGA-PRAD dataset shown here are based upon data generated by the TCGA Research Network: http://cancergenome.nih.gov/. The author would like to acknowledge the U01 CA151261 award that supported collection and sharing of the QIN PROSTATE and QIN-PROSTATE-Repeatability datasets.
Footnote
Conflicts of Interest: Dr. Hulsen is employed by Philips Research. This manuscript assumes that the datasets listed here were collected in a GDPR compliant manner.
References
- American Cancer Society. Key Statistics for Prostate Cancer. Available online: https://www.cancer.org/cancer/prostate-cancer/about/key-statistics.html
- Simell BA, Tornwall OM, Hamalainen I, et al. Transnational access to large prospective cohorts in Europe: Current trends and unmet needs. N Biotechnol 2019;49:98-103. [Crossref] [PubMed]
- New PhD researchers will crunch big data to help fight against prostate cancer. Available online: https://prostatecanceruk.org/about-us/news-and-views/2016/11/new-phd-researchers-will-crunch-big-data-to-help-fight-against-prostate-cancer
- Hulsen T, Jamuar SS, Moody AR, et al. From Big Data to Precision Medicine. Front Med (Lausanne) 2019;6:34. [Crossref] [PubMed]
- Hulsen T, Obbink JH, Van der Linden W, et al. 958 Integrating large datasets for the Movember Global Action Plan on active surveillance for low risk prostate cancer. Eur Urol Suppl 2016;15:e958. [Crossref]
- Hulsen T, Van der Linden W, De Jonge C, et al. PT-073 Developing a future-proof database for the European Randomized study of Screening for Prostate Cancer (ERSPC). Eur Urol Suppl 2019;18:e1766. [Crossref]
- Hulsen T, Obbink H, Schenk E, et al. PCMM Biobank, IT-infrastructure and decision support. CTMM meeting 2013. Available online: http://tim.hulsen.net/documents/pcmm_wp3_130912.pdf
- Hulsen T, Bangma CH. MP70-02 An Overview of Publicly Available Patient-centered Prostate Cancer Datasets. J Urol 2018;199:e934. [Crossref]
- Röthke M, Blondin D, Schlemmer HP, et al. PI-RADS classification: structured reporting for MRI of the prostate. Rofo 2013;185:253-61. [PubMed]
- Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012;2:401-4. [Crossref] [PubMed]
- Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 2002;30:207-10. [Crossref] [PubMed]
- Kolesnikov N, Hastings E, Keays M, et al. ArrayExpress update--simplifying data submissions. Nucleic Acids Res 2015;43:D1113-6. [Crossref] [PubMed]
- Dunning MJ, Vowler SL, Lalonde E, et al. Mining Human Prostate Cancer Datasets: The "camcAPP" Shiny App. EBioMedicine 2017;17:5-6. [Crossref] [PubMed]
- Gandaglia G, Bray F, Cooperberg MR, et al. Prostate Cancer Registries: Current Status and Future Directions. Eur Urol 2016;69:998-1012. [Crossref] [PubMed]
- Gohagan JK, Prorok PC, Kramer BS, et al. Prostate cancer screening in the prostate, lung, colorectal and ovarian cancer screening trial of the National Cancer Institute. J Urol 1994;152:1905-9. [Crossref] [PubMed]
- Surveillance, Epidemiology, and End Results (SEER) Program () Research Data (1973-2015), National Cancer Institute, DCCPS, Surveillance Research Program, released April 2018, based on the November 2017 submission.www.seer.cancer.gov
- 2001–2015 Database: National Program of Cancer Registries and Surveillance, Epidemiology, and End Results SEER*Stat Database: NPCR and SEER Incidence – USCS 2001–2015 Public Use Research Database, United States Department of Health and Human Services, Centers for Disease Control and Prevention and National Cancer Institute. Released June 2018, based on the November 2017 submission. Available online: www.cdc.gov/cancer/uscs/public-use
- 2005–2015 Database: National Program of Cancer Registries and Surveillance, Epidemiology, and End Results SEER*Stat Database: NPCR and SEER Incidence – USCS 2005–2015 Public Use Research Database, United States Department of Health and Human Services, Centers for Disease Control and Prevention and National Cancer Institute. Released June 2018, based on the November 2017 submission. Available online: www.cdc.gov/cancer/uscs/public-use
- Stamey TA, Kabalin JN, McNeal JE, et al. Prostate specific antigen in the diagnosis and treatment of adenocarcinoma of the prostate. II. Radical prostatectomy treated patients. J Urol 1989;141:1076-83. [Crossref] [PubMed]
- Fraser M, Sabelnykova VY, Yamaguchi TN, et al. Genomic hallmarks of localized, non-indolent prostate cancer. Nature 2017;541:359-64. [Crossref] [PubMed]
- Cheng DT, Mitchell TN, Zehir A, et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): A Hybridization Capture-Based Next-Generation Sequencing Clinical Assay for Solid Tumor Molecular Oncology. J Mol Diagn 2015;17:251-64. [Crossref] [PubMed]
- Grasso CS, Wu YM, Robinson DR, et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature 2012;487:239-43. [Crossref] [PubMed]
- Robinson D, Van Allen EM, Wu YM, et al. Integrative clinical genomics of advanced prostate cancer. Cell 2015;161:1215-28. [Crossref] [PubMed]
- Beltran H, Prandi D, Mosquera JM, et al. Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nat Med 2016;22:298-305. [Crossref] [PubMed]
- Baca SC, Prandi D, Lawrence MS, et al. Punctuated evolution of prostate cancer genomes. Cell 2013;153:666-77. [Crossref] [PubMed]
- Barbieri CE, Baca SC, Lawrence MS, et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat Genet 2012;44:685-9. [Crossref] [PubMed]
- Ren S, Wei GH, Liu D, et al. Whole-genome and Transcriptome Sequencing of Prostate Cancer Identify New Genetic Alterations Driving Disease Progression. Eur Urol 2017. [Epub ahead of print]. [PubMed]
- Kumar A, Coleman I, Morrissey C, et al. Substantial interindividual and limited intraindividual genomic diversity among tumors from men with metastatic prostate cancer. Nat Med 2016;22:369-78. [Crossref] [PubMed]
- Taylor BS, Schultz N, Hieronymus H, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell 2010;18:11-22. [Crossref] [PubMed]
- Armenia J, Wankowicz SAM, Liu D, et al. The long tail of oncogenic drivers in prostate cancer. Nat Genet 2018;50:645-51. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. The Molecular Taxonomy of Primary Prostate Cancer. Cell 2015;163:1011-25. [Crossref] [PubMed]
- Hoadley KA, Yau C, Hinoue T, et al. Cell-of-Origin Patterns Dominate the Molecular Classification of 10,000 Tumors from 33 Types of Cancer. Cell 2018;173:291-304.e6. [Crossref] [PubMed]
- Hieronymus H, Schultz N, Gopalan A, et al. Copy number alteration burden predicts prostate cancer relapse. Proc Natl Acad Sci U S A 2014;111:11139-44. [Crossref] [PubMed]
- Gao D, Vela I, Sboner A, et al. Organoid cultures derived from patients with advanced prostate cancer. Cell 2014;159:176-87. [Crossref] [PubMed]
- AACR Project GENIE Consortium. AACR Project GENIE: Powering Precision Medicine through an International Consortium. Cancer Discov 2017;7:818-31. [Crossref] [PubMed]
- Zhang J, Baran J, Cros A, et al. International Cancer Genome Consortium Data Portal--a one-stop shop for cancer genomics data. Database (Oxford) 2011;2011:bar026. [Crossref] [PubMed]
- Bristow R, Boutros P, Hudson T, et al. Prostate Adenocarcinoma - Canada. Available online: https://icgc.org/icgc/cgp/70/392/70542
- Cussenot O. Prostate Adenocarcinoma - France. Available online: https://icgc.org/icgc/cgp/70/355/1002116
- Sültmann H, Sauter G. Early Onset Prostate Cancer - Germany. Available online: https://icgc.org/icgc/cgp/70/345/53039
- Cooper C, Eeles R, Stratton M, et al. Prostate Adenocarcinoma - United Kingdom. Available online: https://icgc.org/icgc/cgp/70/508/71331
- Consortium T. Prostate Adenocarcinoma TCGA - United States. Available online: https://icgc.org/icgc/cgp/70/509/70272
- Sun Y. Prostate Cancer - China. Available online: https://icgc.org/icgc/cgp/70/371/1003238
- Grossman RL, Heath AP, Ferretti V, et al. Toward a Shared Vision for Cancer Genomic Data. N Engl J Med 2016;375:1109-12. [Crossref] [PubMed]
- Clark K, Vendt B, Smith K, et al. The Cancer Imaging Archive (TCIA): maintaining and operating a public information repository. J Digit Imaging 2013;26:1045-57. [Crossref] [PubMed]
- Choyke P, Turkbey B, Pinto P, et al. (2016). Data From PROSTATE-MRI. The Cancer Imaging Archive. Available online: http://doi.org/ [Crossref]
- Bloch BN, Jain A, Jaffe CC (2015). Data From PROSTATE-DIAGNOSIS. The Cancer Imaging Archive. Available online: http://doi.org/ [Crossref]
- Kurdziel KA, Shih JH, Apolo AB, et al. The kinetics and reproducibility of 18F-sodium fluoride for oncology using current PET camera technology. J Nucl Med 2012;53:1175-84. [Crossref] [PubMed]
- Kurdziel KA, Shih JH, Apolo AB, et al. (2015). Data From NaF_PROSTATE. The Cancer Imaging Archive. Available online: http://doi.org/ [Crossref]
- Litjens G, Futterer J, Huisman H (2015). Data From Prostate-3T. The Cancer Imaging Archive. Available online: http://doi.org/ [Crossref]
- Fedorov A, Fluckiger J, Ayers GD, et al. A comparison of two methods for estimating DCE-MRI parameters via individual and cohort based AIFs in prostate cancer: a step towards practical implementation. Magn Reson Imaging 2014;32:321-9. [Crossref] [PubMed]
- Fedorov A, Tempany C, Mulkern R, et al. (2016). Data From QIN PROSTATE. The Cancer Imaging Archive. Available online: http://doi.org/ [Crossref]
- Zuley ML, Jarosz R, Drake BF, et al. (2016). Radiology Data from The Cancer Genome Atlas Prostate Adenocarcinoma [TCGA-PRAD] collection. The Cancer Imaging Archive. Available online: http://doi.org/ [Crossref]
- Madabhushi A, Feldman M (2016). Fused Radiology-Pathology Prostate Dataset. The Cancer Imaging Archive. Available online: http://doi.org/ [Crossref]
- Litjens G, Debats O, Barentsz J, et al. Computer-aided detection of prostate cancer in MRI. IEEE Trans Med Imaging 2014;33:1083-92. [Crossref] [PubMed]
- Litjens G, Debats O, Barentsz J, et al. (2017). ProstateX Challenge data. The Cancer Imaging Archive. Available online: https://doi.org/ [Crossref]
- Fedorov A, Schwier M, Clunie D, et al. An annotated test-retest collection of prostate multiparametric MRI. Sci Data 2018;5:180281. [Crossref] [PubMed]
- Fedorov A, Schwier M, Clunie D, et al. (2018). Data From QIN-PROSTATE-Repeatability. The Cancer Imaging Archive. Available online: http://doi.org/ [Crossref]
- Fedorov A, Vangel MG, Tempany CM, et al. Multiparametric Magnetic Resonance Imaging of the Prostate: Repeatability of Volume and Apparent Diffusion Coefficient Quantification. Invest Radiol 2017;52:538-46. [Crossref] [PubMed]
- Wilkinson MD, Dumontier M, Aalbersberg IJ, et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci Data 2016;3:160018. [Crossref] [PubMed]
- Hulsen T, Van der Linden W, Pletea D, et al. Data Model Mapping. (2017). Available online: https://patentscope.wipo.int/search/en/detail.jsf?docId=WO2017167628
- Darshan M, Zheng Q, Fedor HL, et al. Biobanking of derivatives from radical retropubic and robot-assisted laparoscopic prostatectomy tissues as part of the prostate cancer biorepository network. Prostate 2014;74:61-9. [Crossref] [PubMed]
- Bangma C, Obbink H. The future of prostate cancer research: bringing data together, looking back and forward. Transl Androl Urol 2018;7:188-94. [Crossref] [PubMed]