
Challenges of urine-based molecular assays for the detection of urothelial cancer
In the issue of Modern Pathology, April 25 2019, Eich and colleagues highlight the relevance of a urine assay called ‘UroSEEK’ as a non-invasive approach for the detection of primary or recurrent urothelial carcinoma of the bladder (UCB). The UROSEEK assay consists of three components; TERTSeqS, UroSeqS, and FastSeqS, which cover molecular alterations in 11 genes that frequently occur in UCB, in addition to aneuploidy (FastSeqS) (1). In 2018, the same group reported on the performance of the UroSEEK assay tested in urine samples (2), whereas in the current study, the presence of UroSEEK gene alterations was analyzed in tumor tissue samples. In the current study, it was shown that more than 90% of 527 tissue samples analyzed had at least one mutation in the 11 genes included in the UroSEEK assay.
The rationale behind a urine-based tumor assay is that urothelial tumors located in the urinary tract are in direct contact with urine and shed cells in the urine. Therefore, analyzing cells isolated from urine for the presence of tumor specific features (i.e., morphology, mutations) might indicate the presence of a tumor in the urinary tract. Fact remains that urothelial tumors do not release cells into the urinary tract at all times. Ergo, not all urine samples from patients with a tumor in the urinary tract show presence of urothelial tumor cells. Moreover, tumor cells in urine are present in an (sometimes abundant) background of normal cells. Furthermore, a tumor can be heterogeneous having spatial molecular alterations, which could contribute to the fact that tumor-specific features detected in tissue are not always detectable in cell pellets from urine. To illustrate this, Zuiverloon et al. collected urine samples of 134 patients who had previously been diagnosed with an FGFR3-mutant UCB and were on surveillance (3). The urine samples were analyzed for the presence of an FGFR3 mutation. During follow-up, 45 histologically proven UCB recurrences occurred and in only 26 (58%) the corresponding urine sample tested positive for an FGFR3-mutation. Importantly, FGFR3-negative urine samples were embedded between positive urines preceding a recurrence. In the present study, Eich et al. reported that in 527 tumor tissue samples tested, 487 cases (92%) were positive for at least one of the UroSEEK alterations. Although this percentage represents, as stated by the authors, a ‘comprehensive coverage’, the assay had slightly lower performance when tested in urine samples. In a cohort comprised of patients referred to a urology clinic for hematuria or lower urinary tract symptoms from the previous UroSEEK study (2018, ‘early detection cohort’), the assay had a sensitivity of 83%, with at least one positive UroSEEK marker in 145/175 patients that had a urothelial tumor. Notably, when evaluated in urine samples, detection assays also tend to have a specificity reported as part of their performance, a metric that cannot be assessed when testing assay coverage within tumor tissue. However, for the ‘early detection cohort’, UroSEEK’s specificitiy was reported to be 93% (Table 1).

Full table
Many efforts have been made for the development of non-invasive assays for the detection of UCB, but urine-based tumor markers are not implemented yet in the guidelines for the diagnosis and surveillance of bladder cancer patients. Lotan et al. recently discussed the lack of clarity regarding the clinical benefit of urine-based tumor markers and highlighted that the usefulness of a marker depends on the clinical setting in which it will be used (4); i.e., a primary versus a surveillance setting. According to clinical guidelines, patients presenting with hematuria need to be evaluated by cystoscopy to rule out the presence of a bladder tumor. However, as the prevalence of UCB in patients presenting hematuria varies from 2–28% (5), the majority of cystoscopies are ‘unnecessarily’ executed. Therefore, in the primary diagnostic setting, a urine-based assay could be used to triage hematuria patients for diagnostic cystoscopy. One could argue that in this particular setting a urine marker requires the highest possible sensitivity i.e., negative predictive value to be clinically useful, as preferably, no urothelial cancer patients should be wrongly withheld from a diagnostic cystoscopy. And, although a highly sensitive marker is likely to bring false positive test results, this is considered acceptable, since currently all hematuria patients are scheduled for a cystoscopy.
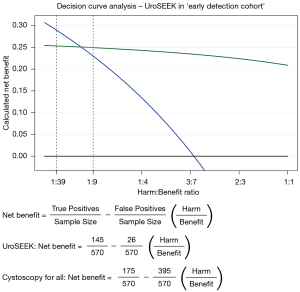
According to van Calster et al. (6), traditional statistical measures, like sensitivity and specificity, do not provide an answer as to whether an assay should be implemented in clinical practice. Importantly, in order to properly evaluate a diagnostic assay, one should conduct a so-called ‘decision curve analysis’. In this analysis, the clinical value of an assay is assessed by incorporating the clinical consequences of using such an assay. In the next paragraph we would like to illustrate this type of analysis, with the example of the reported UroSEEK sensitivity/specificity values in the ‘early detection cohort’ [Springer et al. Table 1 (2)].
For the primary diagnostic setting, a decision curve analysis takes into account that conducting a cystoscopy based on a positive test result, carries ‘benefit’ in urothelial cancer patients (true positives), and that it carries ‘harm’ when conducted in patients that do not appear to have urothelial cancer (false positives). The decision curve analysis is then based on how many diagnostic cystoscopies a urologist is willing to conduct, in order to find one urothelial cancer case.
Suppose a urologist is willing to conduct 40 cystoscopies in order to find 1 urothelial cancer case, the Harm:Benefit ratio would be 1:39. Another urologist may find an unnecessary cystoscopy to be more harmful, stating that he/she would be willing to conduct 10 cystoscopies in order to find 1 urothelial cancer case. In this case, the Harm:Benefit ratio would be 1:9. Consequently, one can calculate the ‘net benefit’ of using their assay to triage patients for cystoscopy (Figure 1). According to van Calster et al. a triage-assay is to be considered for clinical implementation, when the calculated ‘net benefit’ of the assay is higher than current clinical practice (here; cystoscopy for all patients). Figure 1 shows a superior net benefit for the UroSEEK assay at a Harm:Benefit ratio of 1:9, whereas the assay’s net benefit is lower than ‘cystoscopy for all’, at a Harm:Benefit ratio of 1:39.
Whereas the reported coverage of the UroSEEK assay looks promising, one has to be cautious when interpreting case-enriched or retrospective data. As a reason for not being implemented in the clinic yet, Tan et al. stressed in their review that a significant number of developed assays are not externally validated in prospective field testing (7). It is important to realise that the observed assay performance heavily relies on the underlying tumor prevalence of the cohort in which it is evaluated. As also stated by Springer et al., ‘by design, the fraction of cases in the ‘early detection cohort’ was higher than the fraction of patients with similar presentations who would have developed UCB in standard clinical practice’ (2). Furthermore, it is known that tumor prevalence differs between microscopic and gross hematuria populations and one could argue that in gross hematuria patients replacing cystoscopy by a urine assay will not be suitable at all, due to the high a priori chance of a tumor and the need to exclude other causes of hematuria as soon as possible. In the ‘early detection cohort’, patients were not stratified for the type of hematuria. Thirdly, in the real-world clinical setting, the occurrence of low-grade non-invasive papillary carcinoma is higher. In contrast, Eich et al. tested the coverage of the UroSEEK assay on 527 tumors from 484 patients of which the majority of cases (64%) were high-grade/invasive carcinoma. Therefore, an important result of the study is that at least one UroSEEK gene alteration was found in 98% (185/188) of the subset low-grade non-invasive papillary carcinomas (1), suggesting a promising overall sensitivity when evaluated prospectively.
The UroSEEK assay seems to have superior sensitivity to cytology and the sensitivity increases when the two are combined (2). These data suggest that the assay could be a useful tool in the primary diagnostic setting. In order to be clinically useful in the surveillance setting, however, the assay needs a suitable specificity and consequently a high positive predictive value. The reported specificity of the UroSEEK assay was 93% in the ‘early detection cohort’ versus 80% in a surveillance cohort (N=322) with a sensitivity of 68%. As stated by the authors, the proportion of false positives could be related to a lead time ahead of clinical detection (2) [also known as the ‘anticipatory’ effect (3)]; the assay might detect a minimal amount of tumor cells in patients where a tumor is still clinically undetectable. However, a ‘false’ positive test result can cause unnecessary distress in patients, especially when a tumor cannot be detected at all. In context, Lotan et al. explained that because of the high specificity of cytology, biopsies and upper tract imaging are indicated to find the location of a possible recurrence, when cytology shows suspicion of high grade urothelial carcinoma (4). Most studies evaluating the diagnostic accuracy of a urinary assay did not conduct a confirmatory biopsy when their assay tested positive. A high specificity, together with such an evaluation, is mandatory if one ever wants to prove the superiority of an assay over urinary markers that are currently used.
DNA-based urine assays need a certain amount of input DNA in order to yield a positive or negative test result. When evaluating assay coverage on tumor tissue, there is likely enough DNA input for the assay to be performed, but when retrieving DNA from urine samples, not every sample necessarily contains enough DNA. Nonetheless, Springer et al. described that they designed a specific set of primers that allowed to detect mutations in as few as 0.03% of cells in the urine and their simulations demonstrated that DNA containing a minimum of 1% neoplastic cells was required for reliable aneuploidy detection (2). Data was not provided on the proportion of samples that could not be analyzed due to low DNA yield. Furthermore, Eich et al. did not discuss the possible influence of intravesical/neo-adjuvant therapy on the result of the UroSEEK assay, especially within the 36 patients for which tissue from sequential tumors was analyzed (1). Importantly, the authors observed disconcordant mutational patterns in these sequential tumors, which might be a result from the impact of antitumor treatment. When conducting a prospective- or interventional randomized controlled trial, failed test results are more likely to happen, especially in the surveillance setting, where DNA yield is generally lower due to earlier detection of smaller recurrences. Lastly, despite the fact that the UroSEEK target gene selection is based on an extensive literature evaluation (2), the current study observed no MET gene mutations and only one MLL mutation in 527 tumor samples tested (1). Since decreasing the number of markers will likely result in a lower change of false positive test results, when evaluating UroSEEK in urine samples, one could consider to remove these targets.
In conclusion, the authors of the series on the diagnostic performance of the UroSEEK assay in both urine and tissue samples are congratulated for undertaking such large assay-evaluation studies. Although more evidence is required to determine the clinical usefulness of the assay, it is clear that the authors have put a tremendous effort in the development of this assay that has a high reproducibility, a high coverage corresponding with a sensitivity that is consistently higher than cytology, and even has utility in detecting upper tract disease.
Acknowledgments
None.
Footnote
Conflicts of Interest: JJJ has performed consulting activities for Decipher Biosciences. JJJ and KEK received travel grants from MDxHealth to attend scientific meetings. MJR has no conflict of interest to report. JLB has performed consulting activities for MSD, Roche, BMS and Janssen Pharmaceuticals and has a research agreement with Decipher Biosciences.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
References
- Eich ML, Rodriguez Pena MDC, Springer SU, et al. Incidence and distribution of UroSEEK gene panel in a multi-institutional cohort of bladder urothelial carcinoma. Mod Pathol 2019. [Epub ahead of print]. [Crossref] [PubMed]
- Springer SU, Chen CH, Rodriguez Pena MDC, et al. Non-invasive detection of urothelial cancer through the analysis of driver gene mutations and aneuploidy. Elife 2018;7. [Crossref] [PubMed]
- Zuiverloon TC, van der Aa MN, van der Kwast TH, et al. Fibroblast growth factor receptor 3 mutation analysis on voided urine for surveillance of patients with low-grade non-muscle-invasive bladder cancer. Clin Cancer Res 2010;16:3011-8. [Crossref] [PubMed]
- Lotan Y, Black PC, Caba L, et al. Optimal Trial Design for Studying Urinary Markers in Bladder Cancer: A Collaborative Review. Eur Urol Oncol 2018;1:223-30. [Crossref] [PubMed]
- Georgieva MV, Wheeler SB, Erim D, et al. Comparison of the Harms, Advantages, and Costs Associated With Alternative Guidelines for the Evaluation of Hematuria. JAMA Intern Med 2019. [Epub ahead of print].
- Van Calster B, Wynants L, Verbeek JFM, et al. Reporting and Interpreting Decision Curve Analysis: A Guide for Investigators. Eur Urol 2018;74:796-804. [Crossref] [PubMed]
- Tan WS, Tan WP, Tan MY, et al. Novel urinary biomarkers for the detection of bladder cancer: A systematic review. Cancer Treat Rev 2018;69:39-52. [Crossref] [PubMed]


