
Novel method to detect, isolate, and culture prostate culturing circulating tumor cells
Introduction
Recent evidence has indicated that circulating tumor cells (CTCs) play a critical role in cancer metastasis by utilizing the circulatory system (1-4). Thus, CTCs can serve as cancer markers for early metastasis diagnosis and monitoring anti-cancer therapeutic efficacy, and as cancer targets for creating anti-cancer interventions (5-9). At the moment, the FDA approved CTC detection method is based on the antibody binding (such as the CellSearchTM system), which is not yet optimized (10). For example, the dissociation factor of immunoreaction and the variation of antibody preparation from lot to lot makes it difficult to consistently catch a high percentage of CTCs. When dealing with the low frequency of CTCs in the blood, an additional step is required for the CellSearchTM, which involves enriching the epithelial cell adhesion molecule (EpCAM)-positive CTCs using anti-EpCAM antibodies attached to small solids, such as nanoparticles (10). The process of preparing the immunoparticles is time consuming and inefficient due to the loss of CTCs on immunoparticles during the enrichment procedure. Additionally, the specific antibody-bound epitopes on the EpCAM molecule can be changed from CTCs to CTCs. Thus, the single antibody-based detection may have different binding affinities to the CTCs from different patients and under different assay conditions. The antibody-based systems also require several steps in the procedure such as cartridge preparation, blood sample centrifuge, and multiple step washings which increases the chances of losing the caught CTCs.
In general, the isolated CTCs from patient blood can be used for the characterization of individual cancer properties and as drug targets to optimize chemotherapy. However, the antibody-based method, such as CellSearchTM system can only detect the CTCs, and is difficult to use for isolation and culturing of the CTCs in their native conditions after binding to the EpCAM antibody. These limitations have led to the search for additional methods to detect, isolate, and culture CTCs for cancer diagnosis and drug screening. The goal of this study is to isolate cancer stromal cells (11-13) that express large amounts of EpCAM molecules on the cell surface, allowing them to be used as “bait” to catch other circulating cancer cells expressing EpCAM through cell-cell interaction mechanism. To test this hypothesis, we have used prostate cancer stromal cells as a model establishing a novel method able to combine the three (detection, isolate and culture) in one assay for prostate CTCs.
Methods
Materials
A human prostate cancer line (PC3), human normal prostate cell line (RWPE-1), and F-12K medium for culturing the PC3 cell line were purchased from American Type Culture Collection (ATCC, Manassas, VA, USA). Keratinocyte serum-free medium (K-SFM), and 4',6-diamidino-2-phenylindole (DAPI) were purchased from Invitrogen (Grand Island, NY, USA). Mouse anti-CD133, α2β1 integrin, or CD44 antibodies, and rat anti-mouse EpCAM antibody were purchased from eBioscience (San Diego, CA, USA). 96-well plates and 12-well glass bottom plates were purchased from MatTek Corp (Ashland, MA, USA). Fetal bovine serum (FBS), antibiotic, fluorescent color-labeled secondary antibodies, antimycotic, and other general chemicals were purchased from Sigma (St. Louis, MO, USA). Prostate cancer tissue was obtained from Dr. So, which was used for the previous study (14). Only tissue obtained from non-living mice was used. The institutional and national guide for the care and use of laboratory animals was followed.
Cell culture
The cancer tissue was cut into small pieces, digested using collagenase and then cultured in F-12K medium containing 10% FBS, 1% antibiotic, and antimycotic. The control cells, human normal prostate cell line RWPE-1, were cultured in K-SFM medium containing 0.05 mg/mL bovine pituitary extract (BPE), and 5 ng/mL human recombinant epidermal growth factor (EGF). All of the cell culture experiments were performed as described previously (14,15). The single cell culture was performed on 96-well plate using serial dilution.
Western blot analysis
Western blot analysis was performed as described previously (14,15). Briefly, the cultured cells were collected and washed three times using PBS, and then solubilized in SDS-PAGE buffer containing 2% SDS and analyzed by 12% SDS-PAGE. Bands recognized by individual primary antibodies were visualized with horseradish peroxidase-conjugated secondary antibody as described (14,15).
Immunocytochemical staining for identification of CD133, α2β1 integrin, CD44 and EpCAM on the cell surface
The isolated cells were sub-cultured on a plate and incubated with corresponding antibodies of anti-CD133, α2β1 integrin, CD44 or EpCAM for 10 min. After washing them twice using cell culture medium, the cells were immediately incubated with fluorescent color-labeled rabbit anti-mouse or rat antibody for 20 min. After washing (3 times), the stained CD133, α2β1 integrin, CD44 or EpCAM on the cell’s surfaces were observed immediately using a fluorescent microscope (14,15).
Preparation of the EpCAM-rich prostate cancer stromal cells as “bait” to catch (detect) the other cancer cells
The EpCAM-rich prostate cancer stromal cells and control cells were cultured separately in a 12-well glass bottom plate. DAPI (a nuclear staining fluorescent reagent) was added to the cell culture medium overnight. After removing the unbound DAPI from the cultured cells by washing the cells in the wells three times using serum-free medium, the DAPI-stained cells were ready to be used as “bait” to catch other live and unstained cancer cells.
Establishing a single step method using the “bait” cancer stromal cells to catch the mimicked CTCs in the suspension of the cultured cells
To mimic the CTCs, the cultured prostate cancer cells were suspended in medium and added to the above DAPI-stained prostate cancer stromal cells (with nuclear fluorescent signals attached on the plate) for 1–3 h. After 3 rounds of washing to remove the nonspecific binding using the medium, the caught cancer cells (without fluorescent staining) were clearly distinguished from the DAPI-stained cells under fluorescence microscopy.
Establishing a single step method using the “bait” cancer stromal cells to catch the blood CTCs
For determination of the live CTCs, whole blood (0.2 mL) from a recently terminated cancer mouse with late stage prostate cancer was collected in a tube containing citrate solution (<10%). The blood was mixed with 0.5 mL cell culture medium (serum-free), and then immediately added to the live DAPI-stained prostate cancer stromal cells and DAPI-stained control cells in a 12-well plate. After co-culturing for 1 h, the blood and medium were removed and continually washed three times using serum-free medium. The caught CTCs (without fluorescent signals) from the whole blood were observed under fluorescence microscopy.
Isolation and culturing of the caught prostate CTCs
Once the caught prostate CTCs were identified in the particular well under the fluorescent microscope, the cells were gently detached from the DAPI-stained prostate cancer stromal cells by physically shaking them twice, transferred to a tube, and then diluted into a single cell per well in a 96-well plate. The wells containing only one cell per well without DAPI-nuclear staining were marked as isolated single clones of prostate CTCs candidates. The wells containing cell(s) with DAPI-stained “bait” cell(s) were discarded. The marked single prostate CTC was further cultured using the same medium.
Results
Developing methods to isolate and culture the single clone of prostate cancer stromal cells using limited dilution approach
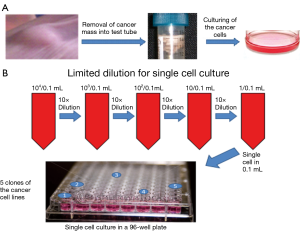
The prostate cancer cells grown from the mouse cancer tissue (Figure 1A) contained a large number of different stages of cancer cells and tiny amount of the stromal cells. continuous multiple step dilutions were used to obtain a single cell per well (called limited dilution, Figure 1B) to isolate cancer stromal cells from non-cancer-stromal. The wells containing a single cell were identified by microscopy. As, a result, out of 96 wells five wells with a single cancer cell able to grow into a cell line were found and named as Clone-1 through 5 of the monoclonal prostate cancer cell lines (Figure 1B).
Identification of the isolated monoclonal prostate cancer as cancer stromal cells
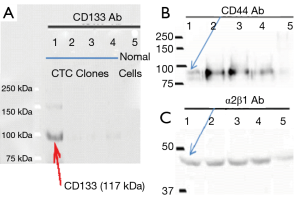
Co-expressing CD133, CD44 and α2β1 intergrin has been used as a marker to identify prostate cancer stromal cells previously (6,7). Thus, the three molecular stains were applied to characterize the Clone 1–5 cell lines as prostate cancer stromal cells. Figure 2 showed that only the Clone-1 cells expressed all three markers (CD133, CD44 and α2β1), as confirmed by cellular immunostaining (Figure 2) and Western blot analysis (Figure 3). Other clones and normal prostate cells were not positive to all of the markers. This indicated that Clone-1 cells are prostate cancer stromal cells.

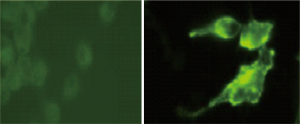
Finding high levels of EpCAM expressed on the surface of the live Clone-1 prostate cancer stromal cells
The Clone-1 cancer stromal cells expressing high levels of EpCAM were observed by immunostaining using an anti-human EpCAM monoclonal antibody (Figure 4 right). In contrast, the normal prostate cells showed negative (very low level) expression of EpCAM (Figure 4 left). This indicated that the Clone-1 cell might be able to specifically attract (catch) other cancer cells including the CTCs, which also express EpCAM molecules because EpCAM-EpCAM interaction is well-known in cancer cell-cell interaction.
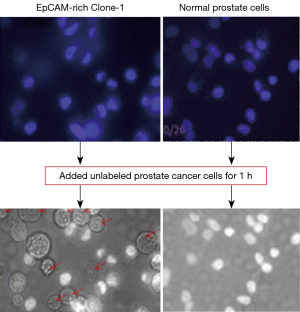
Testing the ability and specificity of the EpCAM-rich Clone-1 cells as “bait” to catch cancer cells through EpCAM-EpCAM of cell-cell interaction principle
Before developing a method to detect CTCs using the EpCAM-rich Clone-1 stromal cells as “bait”, it is important to evaluate its ability and specificity using known cancer cells as standards against the normal cells as control. This is a key step to gaining knowledge on how to make the assay work for CTC detection. In order to clearly distinguish the “bait” cells and the “caught” cancer cells, the EpCAM-rich Clone-1 cells (Figure 5 left) and control cells (normal prostate cells, Figure 5 right) were first labeled with DAPI (nuclear blue stain), and attached to the plate. When the assay started, the cultured prostate cancer cells (as the cancer cell standard) were suspended in medium with a concentration of as little as approximately 1,000 cells per well to mimic CTCs, and then added to the cultured DAPI-“bait” cells (attached on the plate). After as short as a 1 h incubation period, the caught cancer cells by the DAPI-stained “bait” cells were justified by (I) under the normal light microscopy, the caught prostate cancer cells (with a circle shape, without nuclear stain) were identified (Figure 5 bottom left); and (II) under fluorescent microscopy, for the corresponding view, only the DAPI-stained “bait” cells showed blue color nuclear stains, while in contrast the caught cells without DAPI stains were confirmed (Figure 5 top left). It appears that the EpCAM rich Clone-1 stromal cells used as “bait” were able to catch the prostate cancer cells in suspension (Figure 5 left), but the normal prostate cells were not effective as “bait” (Figure 5 right).
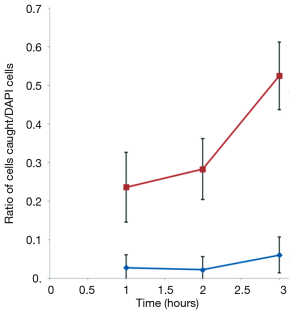
In addition, the DAPI-labeled EpCAM clone-1 cells used as “bait” not binding to the unlabeled normal prostate cells was also confirmed (data not shown). This indicated that the EpCAM-rich Clone-1 cells were able to specifically catch the cancer cells from cell suspension. A time course experiment was further performed to optimize the assay system (Figure 6). Increasing the incubation time to 3 h allowed more prostate cancer cells could be caught by the EpCAM rich Clone-1 stromal cells, but it also increased the background. Thus, a 1-h incubation time was selected for developing an assay for determination of the CTCs (Figure 5).
Comparison of the specificity of the EpCAM-rich Clone-1 cells (left) and normal prostate cells (right) as “bait” to catch cancer cells
Both “bait” cells were labeled by DAPI and cultured in a 12-well plate (all cells were attached on the plate). After the excess DAPI was removed, the live, unlabeled prostate cancer cells were added to the above DAPI-stained “bait” cells. After washing using medium, the cell-cell interaction was observed under fluorescent light, which showed only the DAPI-stained “bait” cells (top panels), and under normal light which were able to show the “bait” cells (white nuclear images) and the caught cancer cells (circular shape without nuclear stain, marked with red arrows (left bottom panel).
Creating a novel cell-cell interaction method to detect CTCs
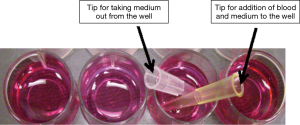
Based on the information from the above study, a simple assay device built with 12-wells plate coated with the DAPI-stained Clone-1 cells along with a convenient use of two pipette tips for adding and washing blood samples and medium was designed (Figure 7). The advantages of the simple assay system are the low cost ($1–3/sample), non-special instrument required, and that it is suitable for any clinic and laboratories with a cell culture environment. The entire procedure for the detection of the CTCs in the blood included the following simple steps: (I) a freshly collected blood sample was injected into the well from the yellow tip and incubated with the DAPI-stained EpCAM-rich Clone-1 cells (which were already attached on the wells, Figure 7); (II) after 1 h, the cells were washed three times by pipetting out the blood and medium from the white tip (Figure 7); (III) the cell-cell interaction was observed under a fluorescence microscope using fluorescent light to mark the DAPI-stained Clone-1 “bait” cells and normal light to identify the non-fluorescent stained CTCs attached to the “bait” cells. Normal prostate cells were used as the negative control for the experiments.
Testing the new assay to detect the blood CTCs of prostate cancer model in mice
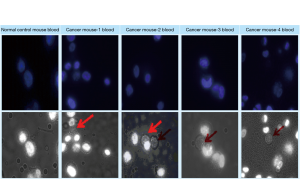
Four of the blood samples [0.2 mL/each, kindly provided from previous study (14)] were collected from each of the later stage prostate cancer mice with visible metastasis (tumor on multiple sites as described previously (14) and subjected to the determination of CTCs using the assay device described above (Figure 7). Using the simple assay device and procedures as described above, the DAPI-stained EpCAM Clone-1 cells used as “bait” were able to catch the CTCs in all of the four blood samples (Figure 8). Similar to that described in Figure 5, the caught CTCs were justified by showing a circular shape without a nuclear stain under normal light (Figure 8 bottom panels) and not showing staining under fluorescent light which was used to show the “bait” cells only (Figure 8 top panels). In contrast, the DAPI-stained EpCAM Clone-1 used as “bait” did not catch any cells when normal blood was used (Figure 8 first column). This experiment has provided strong evidence that the new method using cell-cell interaction is suitable for blood CTC detection.
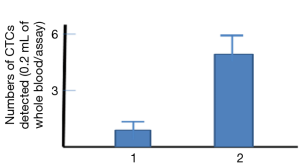
To compare the sensitivity between the new cell-cell interaction-based assay and the old antibody-based assay, similar blood samples used for the above assay (Figure 8) were added to the anti-EPCAM antibody-coated plate; the antibody-based assay only caught one CTC on average (Figure 9 column 1), but the new assay was able to catch five CTCs (Figure 9 column 2). This result indicated that the developed method based on the cell-cell interaction, suitable for detection of CTCs in blood, could be more sensitive than that of antibody-based assays.
Characterizing the novel method as having additional benefits in isolation and culture of the intact CTCs from blood for potential cancer cell cloning and target-based drug screening
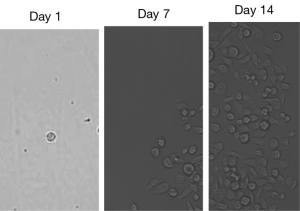
A simple procedure to isolate and culture the caught blood CTCs was developed. The detected CTCs associated with the “bait” cells (EpCAM-rich Clone-1 cells) in the designed device (wells) were under sterilized conditions and within the cell culture medium during the assay. Thus, the CTCs were under live conditions and could be easily dissociated from the “bait” cells through gentle shaking and single cell dilution (limited dilution) as described in Figure 1. After placing the diluted single cell/well to the 96-well place, the single cell without DAPI fluorescent staining identified as the caught CTC was easily distinguished from the DAPI-stained “bait” cell under microscopic analysis using fluorescent light and normal light. Figure 10 shows a single CTC (Figure 10, day 1) isolated by the above dilution method quickly growing into a group of CTCs after 1 week (Figure 10, day 7), and then becoming a monoclonal CTC cell line after 2 weeks (Figure 10, day 14). Thus far, six CTC cell lines have been generated, and stored, which could be used for cancer cloning, gene typing, and anti-prostate cancer drug screening in the future.
Discussion
In this study, we have successfully isolated, cultured, and identified a Clone-1 cell line as a prostate cancer stromal cell line expressing high levels of the surface EpCAM molecule (Figure 4). This molecule has the ability to specifically attract other cancer cells, which also express EpCAM, but not normal cells which are negative in EpCAM expression. Cancer stromal cells are the seed cells that cause cancer recurrence and metastasis (15-17). Thus, cancer stromal cells have more invasive properties when interacting with other tissue and cancer cells (15,16,18). The finding that the EpCAM-positive Clone-1 stromal cells could be used as “bait” to catch trace numbers of prostate cancer cells in suspension and in mimicked circulating blood is the first to the field, and is the basis for the design of the blood CTC detection method. It should be indicated that most cancer CTCs express EpCAM on the surface of the cells. The commercial CTC detection kit is also developed based on the use of anti-EpCAM antibody (18).
We have also established a much simpler, easier, and more sensitive method for blood CTC detection using the cell-cell interaction principle. The addition of the blood sample to the DAPI-stained Clone-1 stromal cell coated plate and then observation under the microscope is a single step. The detection needs only 1-h incubation plus a brief washing. It is much simpler and easier to perform, as other instruments and extra steps such as centrifugation, immunoreactions, and time consuming, expensive analyzing, which are required when use the antibody-based CTC detection methods (18), are unnecessary. Our pilot study for the new cell-based CTC detection is more sensitive than that of the traditional antibody-based CTC assay.
It shall be indicated that the cancer model used for previous study (14,19) was created by a routine method through implantation of the human prostate cancer cell line (with authentication from ATCC) on the back of the immune deficient mice (with authentication from USA Jackson Lab) subcutaneously. The prostate cancer cells growth on the back of the mice were isolated and identified by cancer stem cell markers (Figure 3) and then used for developing of the CTC detection methods. This is a common method for developing different cancer models on immune deficient mice (14,19).
In the developed assay, we used several steps to eliminate the possible contamination of the white blood cells (WBC), which included short time incubation to avoid the adhesion of WBC on the plates; extensive washing to remove any WBC’s remaining on the plate, and >5-day culture to eliminate the survival of the WBC’s. Our controls using normal blood have shown that the WBC contamination was eliminated during the detection and culture (Figure 8).
Another attractive benefit of generating CTC cell lines using the new method is its time efficiency. It is commonly acknowledged that two of many key factors of improving cancer survival rates are speed and early detection. Thus, it is important to identify early cancer metastasis through fast CTC detection. An efficiently developed individual CTC cell line for quick drug screening is necessary to quickly eliminate cancer cells. This method offers CTC detection, isolation, and instant start to culture all in the same day. The CTC cell lines could be generated as targets for drug screening in as quickly as one to 2 weeks.
Scientific logic is important for new research and findings. The developed method was based on the fundamental principle of EpCAM to EpCAM interaction and cancer cell to cancer cell interaction. This principle has been well-documented (20), providing a foundation of scientific mechanisms supporting the developed method. Many patient cancer cell line cultures use cancer masses from biopsy. The noninvasive CTC detection, isolation, and culture three-in-one method could be used prior to biopsy if biopsy is necessary, or serve as an alternative for isolating cancer cells from patients beside the biopsy. Finally, we have also successfully identified additional benefits of the developed method, absent in antibody-based CTC detection assays (such as CellSearchTM) of both isolating and culturing single clone cell of detected CTCs to form individual cell lines combined with the detection assay to create an efficient three-in-one system. Thus, this system has potential to be developed into a high-performance CTC detection, isolation, and culturing system suitable for handling large numbers of samples. The current development has provided a way to easily isolate the cultured CTCs for further characterization. For example, whether other molecules [such as vimentin with cytokeratin (CK), which are expressed in mesenchymal cells] specifically expressed in the prostate CTCs could be characterized using the cultured CTCs. These could be one of our future studies to bring up different topics and discoveries.
Acknowledgments
Funding: This work was supported, in whole or in part, by I.I Rabi Scholars award (for Diana Ruan), Columbia College, and Dean’s award of Vagelos College of Physician and Surgeons (for Diana Ruan), Columbia University, New York, NY, USA; and Cecil M. Crigler MD Chair fund (for Run Wang, MD), Division of Urology, University of Texas Health Science Center-McGovern Medical School at Houston, Department of Surgery, Houston, TX, USA. We thank Dr. Christopher Frey in Baylor College of Medicine providing comments for editing of the manuscript.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study used animal tissue for the experiments, which was approved by the Institutional Animal Care and Use Committee of University of Houston (protocol No: 10-033).
References
- Seyfried TN, Huysentruyt LC. On the origin of cancer metastasis. Crit Rev Oncog 2013;18:43-73. [Crossref] [PubMed]
- Harouaka R, Kang Z, Zheng SY, et al. Circulating tumor cells: advances in isolation and analysis, and challenges for clinical applications. Pharmacol Ther 2014;141:209-21. [Crossref] [PubMed]
- Fidler IJ. The pathogenesis of cancer metastasis: the 'seed and soil' hypothesis revisited. Nat Rev Cancer 2003;3:453-8. [Crossref] [PubMed]
- Ashworth TR. A case of cancer in which cells similar to those in the tumours were seen in the blood after death. Aust Med J 1869;14:146-7.
- Wang Y, Zhou Y, Hu Z. The functions of circulating tumor cells in early diagnosis and surveillance during cancer advancement. J Transl Int Med 2017;5:135-8. [Crossref] [PubMed]
- Yoneda K, Hashimoto M, Takuwa T, et al. Circulating tumor cell (CTC) as a significant prognostic factor in resected primary lung cancer. J Clin Oncol 2017. [Crossref]
- Potdar PD, Lotey NK. Role of circulating tumor cells in future diagnosis and therapy of cancer. J Cancer Metastasis Treat 2015;1:44-56. [Crossref]
- Miller MC, Doyle GV, Terstappen LW. Significance of circulating tumor cells detected by the CellSearch system in patients with metastatic breast colorectal and prostate cancer. J Oncol 2010;2010:617421. [PubMed]
- de Bono JS, Scher HI, Montgomery RB, et al. Circulating tumor cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer. Clin Cancer Res 2008;14:6302-9. [Crossref] [PubMed]
- Veridex CellSearch. 2010. Available online: https://www.cellsearchctc.com
- Lang SH, Frame FM, Collins AT. Prostate cancer stem cells. J Pathol 2009;217:299-306. [Crossref] [PubMed]
- Li C, Heidt DG, Dalerba P, et al. Identification of pancreatic cancer stem cells. Cancer Res 2007;67:1030-7. [Crossref] [PubMed]
- Maitland NJ, Collins AT. Prostate cancer stem cells: a new target for therapy. J Clin Oncol 2008;26:2862-70. [Crossref] [PubMed]
- Chillar A, So SP, Tang N, et al. Identification of tumorigenesis from the specific coupling of cyclooxygenase-2 with microsomal prostaglandin E2 synthase-1 in vivo. Eur J 2014;12:77-88.
- Gupta PB, Chaffer CL, Weinberg RA. Cancer stem cells: mirage or reality? Nat Med 2009;15:1010-2. [Crossref] [PubMed]
- Singh SK, Clarke ID, Terasaki M, et al. Identification of a cancer stem cell in human brain tumors. Cancer Res 2003;63:5821-8. [PubMed]
- Al-Hajj M, Wicha MS, Benito-Hernandez A, et al. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A 2003;100:3983-8. [Crossref] [PubMed]
- CellSearchTM. Raritan, NJ: Veridex, LLC, a Johnson & Johnson company. 2013.
- Ruan D, So SP. Prostaglandin E2 produced by inducible COX-2 and mPGES-1 promoting cancer cell proliferation in vitro and in vivo. Life Sci 2014;116:43-50. [Crossref] [PubMed]
- Ni J, Cozzi PJ, Duan W, et al. Role of the EpCAM (CD326) in prostate cancer metastasis and progression. Cancer Metastasis Rev 2012;31:779-91. [Crossref] [PubMed]


