
Integrative analysis of ceRNA network and DNA methylation associated with gene expression in malignant pheochromocytomas: a study based on The Cancer Genome Atlas
Introduction
Primarily arising from the chromaffin cells of the adrenal medulla (80–85%) or extra-adrenal paraganglia (15–20%) (1), pheochromocytomas (PCCs) are rare catecholamine-secreting tumors with a highly variable clinical presentation, including hypertension, headache, sweating, and palpitation (2,3). Approximately 10% of patients with PCC are malignant, which are defined by distant metastases, and the differential diagnosis between malignant and benign tumors remains challenging. Moreover, the risk of malignancy greatly exceeds the classical 10% in patients with extra-adrenal disease—the so called paragangliomas (1,4). Individuals with malignant PCCs have unfavorable 5 year survival rates that vary between 20% and 50% (5).
In the past decade, major advances in the understanding of the pathogenesis and methods for diagnosing malignant PCCs have been reported. In messenger RNA (mRNA) expression array data, contactin 4 (CNTN4) is more frequently expressed in malignant PCCs than in benign PCCs, and CNTN4 expression is consistently associated with malignant behavior in PCCs (6). In addition, differentially expressed microRNAs (miRNAs) in benign and malignant PCCs have been characterized and found to be markers of malignancy or disease recurrence (7). Recently, long noncoding RNAs (lncRNAs) hold great promise as novel biomarkers and therapeutic targets for various types of cancer because of their genome-wide expression patterns and tissue-specific expression characteristics (8). LncRNA-mediated gene expression involves various mechanisms, such as transcription regulation, translation, protein modification, and cell signaling pathways. However, few studies have directly explored the mechanisms of lncRNAs in malignant PCCs.
The abnormal expression of competitive endogenous RNAs (ceRNAs) is closely related to the occurrence, development, and prognosis of tumors (9). CeRNAs refer to transcripts, such as mRNA, tRNA, rRNA, lncRNA, pseudogene RNA, and circular RNA (10). CeRNAs act as molecular sponges for miRNA through miRNA response elements (MREs) and form complex regulatory networks (11). As ceRNAs, lncRNAs serve as miRNA sponges, playing important roles in the regulation of cancer-related genes. For example, lnc-MD1, a muscle-specific long noncoding RNA, governs the time of muscle differentiation by acting as a ceRNA in mouse and human myoblasts. Lnc-MD1 sponges miR-133 to regulate the expression of MAML1 and MEF2C, which are transcription factors that activate muscle-specific gene expression (12). Moreover, lnc-HULC acts as an endogenous sponge that downregulates a series of miRNA activities, including miR-372. MiR-372 inhibition reduces the translational repression of its target gene, PRKACB, which in turn induces CREB phosphorylation (13). In addition, the association of the RNA-binding protein HuR with lnc-p21 favors the recruitment of let-7/Ago2 to lnc-p21, and thus lnc-p21 stability is low (14). DNA methylation plays an important role in the regulation of gene expression through an epigenetic control in various biological processes and diseases (15). In recent years, the DNA methylation of specific CpG islands has been detected in many types of malignant tumors, and DNA methylation aberrancies are considered hallmarks of human cancers (16). Therefore, an integrative analysis of ceRNAs and DNA methylation associated with gene expression should be conducted to provide novel clues to the pathogenesis of malignant PCCs.
In the present study, we first investigated potential differentially expressed lncRNAs, miRNAs, and mRNAs in malignant PCCs on the basis of The Cancer Genome Atlas (TCGA) database. Subsequently, we constructed a differentially expressed lncRNA-miRNA-mRNA ceRNA network and performed methylation analysis and overall survival analysis on the components of the network to identify malignant PCC-related prognostic biomarkers. This study provided potential lncRNA, miRNA, and mRNA biomarkers, which are expected to contribute to the early diagnosis, treatment, and prognosis of malignant PCCs.
Methods
Analysis of differentially expressed genes on TCGA
The relevant data provided by TCGA are publicly available and open ended and do not require the approval of the local ethics committee (17). All data were derived from the TCGA database (https://tcga-data.nci.nih.gov/), including clinical sample information, mRNA-Seq, miRNA-seq, and lncRNA-seq data, from malignant PCCs. All data were acquired using the Illumina HiSeq 2000 RNA-Seq platform. Sequencing data were converted through Homo_sapiens. GRCh38.94.chr.gtf, which was downloaded from the Ensembl database (https://asia.ensembl.org/index.html). Basing on the Empirical Analysis of Digital Gene Expression Data package (edgeR) and the “limma” package in R, we normalized and analyzed the downloaded data to obtain differentially expressed mRNA, miRNA, and lncRNA molecules. The cut-off value of |log2 fold Change (FC)| was >2.0, and the adjusted P value was <0.01.
Prediction of lncRNA-miRNA and miRNA-mRNA interactions
In this study, we used miRcode (http://www.mircode.org/) to analyze the interactions between lncRNAs and miRNAs on the basis of the differential expression of malignant PCC-specific miRNAs. We used miRTarBase (http://mirtarbase.mbc.nctu.edu.tw), miRDB (http://www.mirdb.org/miRDB), and TargetScan (http://www.targetscan.org) online analysis tools to predict miRNA-targeted mRNAs. The mRNAs that appeared in at least 2 of 3 online prediction tools were selected to overlap with the differentially expressed mRNAs in malignant PCCs. The intersection between two groups were treated as the target mRNA.
Construction of the lncRNA/mRNA/miRNA ceRNA network
Basing on the differentially expressed lncRNAs, miRNAs, and mRNAs and the lncRNA-miRNA and miRNA-mRNA pairs, we used Cytoscape v3.6.1 to establish the lncRNA-miRNA-mRNA ceRNA network. Then, we employed the Database for Annotation, Visualization, and Integrated Discovery to obtain information for Gene Ontology (GO) biological processes. We used a Cytoscape plug-in (BNGO) tool, which involves the following three parts: biological process (BP), cellular component (CC), and molecular function (MF) to further understand the function of these differentially expressed genes in malignant PCCs. The selection criteria were set as an adjusted P<0.05 after FDR correction.
Methylation analysis of differentially expressed genes
DNA methylation data were downloaded from TCGA database and estimated by the beta value. As previously mentioned, the limma package in R was used to obtain differential methylation genes (DMGs) with a cut-off value of |log2FC| >1.0 and P value of <0.05 (18). An overlap was conducted between DMGs and differentially expressed genes (lncRNA, miRNA, mRNA) involved in ceRNA network. The Pearson correlation coefficient was calculated between the expression and methylation level of overlapping genes. Moreover, to further explore the relationship between gene expression and methylation, we searched the methylation sites that were negatively correlated with gene expression.
Relationship of differentially expressed genes with prognosis
The clinical data of patients, including prognosis and survival time, were obtained from TCGA. Survival packages in R and Kaplan-Meier curves were used to assess the relationship between the expression of differentially expressed lncRNAs, miRNAs, and mRNAs and overall survival in patients with malignant PCCs. A P value of <0.05 was considered statistically significant.
Results
Differential RNA expression analysis based on TCGA
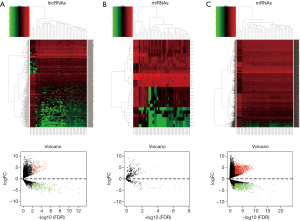
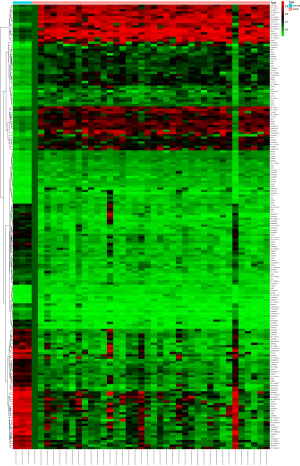
We extracted and analyzed expression profiles of lncRNA, miRNA, and mRNA between 38 malignant PCCs and three control samples on TCGA. Heat maps on all linkage clustering of differentially expressed RNAs along with volcano plots were constructed by R (Figure 1). In total, 447 differentially expressed lncRNAs and 26 differentially expressed miRNAs were screened out. Among the differentially expressed lncRNAs, 286 were downregulated, and 161 were upregulated. Differentially expressed miRNAs contained 21 downregulated miRNAs and five upregulated miRNAs. Moreover, 1,607 differentially expressed mRNAs were found, of which 660 were downregulated and 947 were upregulated.
LncRNA-miRNA and miRNA-mRNA interactions
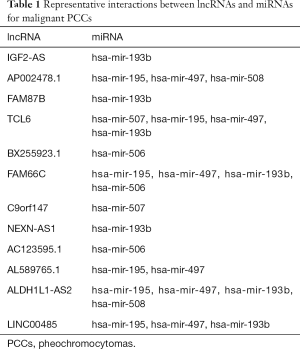
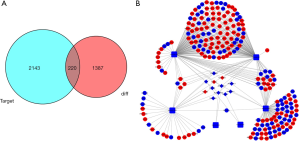
Using the miRcode tool, we compared differentially expressed lncRNA-miRNA pairs and compared 12 differentially expressed lncRNAs and six differentially expressed miRNAs (Table 1). Subsequently, the target genes of the six miRNAs were predicted by miRTarBase, miRDB, and TargetScan online tools. Finally, 2,363 target mRNAs were identified. Furthermore, to obtain a differentially expressed miRNA-mRNA interaction pair in malignant PCCs, we matched the predicted target gene with differentially expressed mRNAs (Figure 2A). MiRNA-targeted mRNAs were excluded if they were not included in differentially expressed mRNAs. A total of 26 differentially expressed lncRNA-miRNA pairs and 366 differentially expressed miRNA-mRNA interaction pairs were obtained (Tables 1 and 2).
Full table

Full table
Construction of ceRNA network in malignant PCCs
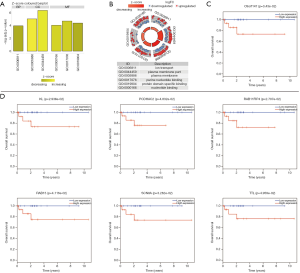
To further investigate the molecular mechanism of malignant PCCs, we constructed a ceRNA network on the basis of the interactions of 26 lncRNA-miRNA and 366 miRNA-mRNA pairs. Finally, 12 lncRNAs, 6 miRNAs, and 220 mRNAs were included in the ceRNA network (Figure 2B). Subsequently, we performed functional enrichment analysis of these differentially expressed mRNAs using the BNGO tool. Top six GO terms were identified in which differentially expressed mRNAs were greatly enriched in BP, CC, and MF (Figures 3A,B). Ion transport was the most significant GO term of BP, whereas plasma membrane was largely enriched in CC. For MF, purine nucleotide binding, protein domain specific binding, and nucleotide binding were the three most relevant terms.
Survival analysis of vital differentially expressed RNAs in malignant PCCs
Kaplan-Meier curves analysis was used in analyzing the correlation of differentially expressed genes in the ceRNA network to overall survival. We found that seven differentially expressed genes were considered to play important roles in the survival outcome of malignant PCCs. LncRNA C9orf147 expression was significantly associated with poor overall survival (log-rank P=0.0343, Figure 3C). Similar to the lncRNA, high expressions of six mRNAs (Klotho, PCDHAC2, RAB11FIP4, RAB15, SCN8A, TTL) significantly predict poor survival outcome (Figure 3D).
Cross-analysis of gene expression and DNA methylation profiles
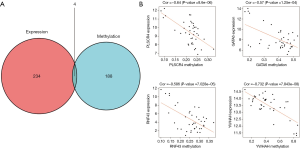
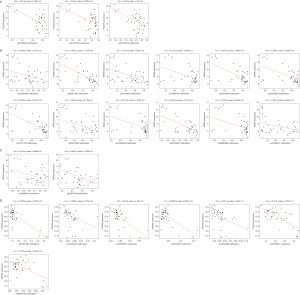
A total of 192 differentially methylated genes were obtained from patients with malignant PCCs and compared with those of the controls (Figure 4). Four of these differentially expressed mRNAs from ceRNAs overlapped with the methylation data (PLSCR4, GATA6, YWHAH, and RNF43). The Pearson's rank test for the genes showed significantly negative correlation between their expression and methylation levels (Figure 5). Of the four genes, three (PLSCR4, GATA6, RNF43) were downregulated and hypermethylated and one (YWHAH) was upregulated and hypomethylated in tumor tissues. Moreover, the methylation sites of these four genes negatively correlated with expression level were sought (Figure S1).
Discussion
Predicting and managing the malignancy of PCCs remain as a considerable challenge in clinical practice (19). Thus, further understanding of the clinical and molecular characteristics of patients with malignant PCCs is urgently needed. In the present study, we performed a bioinformatics analysis on the differentially expressed lncRNAs, miRNAs, and mRNAs in malignant PCCs and compared them with nontumor tissues based on the TCGA database. We further analyzed the function of malignant PCCs-specific mRNAs by using the GO biological analysis. The GO analysis revealed that the functions of these genes were mainly enriched in the regulation of ion transport, plasma membrane, and nucleotide and protein domain-specific binding.
On the basis of the ceRNA network in malignant PCCs, we analyzed the potential mechanism of interaction among lncRNAs, miRNAs, and mRNAs. Of the 12 lncRNAs identified in the ceRNA network, high lnc-C9orf147 expression was significantly related to poor overall survival. According to our ceRNA network, lnc-C9orf147 is involved in the interaction with miR-507, which is a tumor suppressor in many cancer cells (20). However, the involvement of lnc-C9orf147 in other diseases or malignancies and its function have not been reported. Thus, we considered lnc-C9orf147 as a promising biomarker that needs further investigation and definition.
In addition, six differentially expressed mRNAs from the ceRNA network were significantly associated with the prognosis of malignant PCCs. One of the six mRNAs is Klotho (KL), which produces two types of proteins: α-Klotho and β-Klotho. These proteins are the essential components of endocrine fibroblast growth factor (FGF) receptor complexes. The FGF-KL endocrine system has a crucial role in the pathophysiology of aging-related disorders, including diabetes, cancer, arteriosclerosis, and chronic kidney disease (21). KL is dysregulated in several cancers, and its downregulation is related to the promoted proliferation and reduced apoptosis of tumor cells (22). Furthermore, the KL protein provides new insights into cancer target treatment. KL overexpression has synergistic effects with cisplatin to inhibit the proliferation of drug-resistant lung cancer cells in a dose-and time-dependent manner (23). Furthermore, KL overexpression inhibits the progression of ovarian cancer in nude mice partly via the inhibition of systemic inflammation (24). However, our result indicated that KL was upregulated in malignant PCCs and was significantly related to poor overall survival, which differed from other mentioned cancers. The upregulation of SCN8A-mediated invasiveness of cervical cancer cells involves MMP-2 activity, and SCN8A channels are considered therapeutic targets against cancer metastasis (25,26). Few studies have investigated the function and prognostic significance of the other four identified mRNAs.
Regarding the methylation profile analysis associated with gene expression in the ceRNA network, four mRNAs, namely, PLSCR4, GATA6, YWHAH, and RNF43, were identified. PLSCR4, GATA6, and RNF43 were downregulated and hypermethylated, and YWHAH was upregulated and hypomethylated in malignant PCCs. The expression of GATA6 was epigenetically silenced through promoter methylation in gastric cancer cell lines, and aberrant JAK/STAT signaling suppresses TFF1/2 partially through the epigenetic silencing of GATA6. Moreover, gastric cancer patients with high GATA6 methylation tend to have short overall survival (27). The promoter hypermethylation of GATA6 has high frequency in glioblastoma and is an independent predictor in glioblastoma patient outcome (28,29). GATA6 is also hypermethylated in both rhabdomyosarcoma cell lines and primary samples (30). RNF43 is a significantly mutated driver gene in gastric cancer. However, no relevant studies have focused on the gene methylation of PLSCR4 or YWHAH in human cancers and malignancies.
Although this study has shown outcomes with significant clinical significance, we still need to consider several limitations. First, the sample number of the control group from TCGA database was relatively small. Thus, more cases need to be enrolled for analysis. Second, the molecular mechanisms and prognostic roles of lnc-C9orf147 and vital methylated genes in malignant PCCs needs to be further studied in vivo and in vitro.
Conclusions
We constructed a ceRNA network with malignant PCC-specific lncRNAs, miRNAs, and mRNAs in this study. We clarified the mechanism of DNA methylation-associated genes in a ceRNA network in malignant PCCs. Our research increases the understanding of the pathogenesis of malignant PCCs and offers potential genes as underlying therapeutic targets or prognostic biomarkers.
Acknowledgments
Funding: None.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tau.2020.01.29). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The relevant data provided by TCGA are publicly available and open-ended, and do not require the approval of the local ethics committee.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Elder EE, Elder G, Larsson C. Pheochromocytoma and functional paraganglioma syndrome: no longer the 10% tumor. J Surg Oncol 2005;89:193-201. [Crossref] [PubMed]
- Lenders JW, Eisenhofer G, Mannelli M, et al. Phaeochromocytoma. Lancet 2005;366:665-75. [Crossref] [PubMed]
- Harari A, Inabnet WB 3rd. Malignant pheochromocytoma: a review. Am J Surg 2011;201:700-8. [Crossref] [PubMed]
- Strong VE, Kennedy T, Al-Ahmadie H, et al. Prognostic indicators of malignancy in adrenal pheochromocytomas: clinical, histopathologic, and cell cycle/apoptosis gene expression analysis. Surgery 2008;143:759-68. [Crossref] [PubMed]
- Gimm O, DeMicco C, Perren A, et al. Malignant pheochromocytomas and paragangliomas: a diagnostic challenge. Langenbecks Arch Surg 2012;397:155-77. [Crossref] [PubMed]
- Evenepoel L, van Nederveen FH, Oudijk L, et al. Expression of Contactin 4 Is Associated With Malignant Behavior in Pheochromocytomas and Paragangliomas. J Clin Endocrinol Metab 2018;103:46-55. [Crossref] [PubMed]
- Igaz P, Igaz I, Nagy Z, et al. MicroRNAs in adrenal tumors: relevance for pathogenesis, diagnosis, and therapy. Cell Mol Life Sci 2015;72:417-28. [Crossref] [PubMed]
- Bhan A, Soleimani M, Mandal SS. Long Noncoding RNA and Cancer: A New Paradigm. Cancer Res 2017;77:3965-81. [Crossref] [PubMed]
- Qi X, Zhang DH, Wu N, et al. ceRNA in cancer: possible functions and clinical implications. J Med Genet 2015;52:710-8. [Crossref] [PubMed]
- Qu S, Yang X, Li X, et al. Circular RNA: A new star of noncoding RNAs. Cancer Lett 2015;365:141-8. [Crossref] [PubMed]
- Xu J, Li Y, Lu J, et al. The mRNA related ceRNA-ceRNA landscape and significance across 20 major cancer types. Nucleic Acids Res 2015;43:8169-82. [Crossref] [PubMed]
- Cesana M, Cacchiarelli D, Legnini I, et al. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell 2011;147:358-69. [Crossref] [PubMed]
- Wang J, Liu X, Wu H, et al. CREB up-regulates long non-coding RNA, HULC expression through interaction with microRNA-372 in liver cancer. Nucleic Acids Res 2010;38:5366-83. [Crossref] [PubMed]
- Yoon JH, Abdelmohsen K, Srikantan S, et al. LincRNA-p21 suppresses target mRNA translation. Mol Cell 2012;47:648-55. [Crossref] [PubMed]
- Li E, Zhang Y. DNA methylation in mammals. Cold Spring Harb Perspect Biol 2014;6:a019133. [Crossref] [PubMed]
- Liang G, Weisenberger DJ. DNA methylation aberrancies as a guide for surveillance and treatment of human cancers. Epigenetics 2017;12:416-32. [Crossref] [PubMed]
- Gao C, Li H, Zhuang J, et al. The construction and analysis of ceRNA networks in invasive breast cancer: a study based on The Cancer Genome Atlas. Cancer Manag Res 2018;11:1-11. [Crossref] [PubMed]
- Li Z, Zhang R, Yang X, et al. Analysis of gene expression and methylation datasets identified ADAMTS9, FKBP5, and PFKBF3 as biomarkers for osteoarthritis. J Cell Physiol 2019;234:8908-17. [Crossref] [PubMed]
- Roman-Gonzalez A, Jimenez C. Malignant pheochromocytoma-paraganglioma: pathogenesis, TNM staging, and current clinical trials. Curr Opin Endocrinol Diabetes Obes 2017;24:174-83. [Crossref] [PubMed]
- Yamamoto S, Inoue J, Kawano T, et al. The impact of miRNA-based molecular diagnostics and treatment of NRF2-stabilized tumors. Mol Cancer Res 2014;12:58-68. [Crossref] [PubMed]
- Kuro-O M. The Klotho proteins in health and disease. Nat Rev Nephrol 2019;15:27-44. [Crossref] [PubMed]
- Zhou X, Wang X. Klotho: a novel biomarker for cancer. J Cancer Res Clin Oncol 2015;141:961-9. [Crossref] [PubMed]
- Chen T, Ren H, Thakur A, et al. Decreased Level of Klotho Contributes to Drug Resistance in Lung Cancer Cells: Involving in Klotho-Mediated Cell Autophagy. DNA Cell Biol 2016;35:751-7. [Crossref] [PubMed]
- Yan Y, Wang Y, Xiong Y, et al. Reduced Klotho expression contributes to poor survival rates in human patients with ovarian cancer, and overexpression of Klotho inhibits the progression of ovarian cancer partly via the inhibition of systemic inflammation in nude mice. Mol Med Rep 2017;15:1777-85. [Crossref] [PubMed]
- Lopez-Charcas O, Espinosa AM, Alfaro A, et al. The invasiveness of human cervical cancer associated to the function of NaV1.6 channels is mediated by MMP-2 activity. Sci Rep 2018;8:12995. [Crossref] [PubMed]
- Hernandez-Plata E, Ortiz CS, Marquina-Castillo B, et al. Overexpression of NaV 1.6 channels is associated with the invasion capacity of human cervical cancer. Int J Cancer 2012;130:2013-23. [Crossref] [PubMed]
- Wu CS, Wei KL, Chou JL, et al. Aberrant JAK/STAT Signaling Suppresses TFF1 and TFF2 through Epigenetic Silencing of GATA6 in Gastric Cancer. Int J Mol Sci 2016;17. [Crossref] [PubMed]
- Cecener G, Tunca B, Egeli U, et al. The promoter hypermethylation status of GATA6, MGMT, and FHIT in glioblastoma. Cell Mol Neurobiol 2012;32:237-44. [Crossref] [PubMed]
- Skiriute D, Vaitkiene P, Saferis V, et al. MGMT, GATA6, CD81, DR4, and CASP8 gene promoter methylation in glioblastoma. BMC Cancer 2012;12:218. [Crossref] [PubMed]
- Mahoney SE, Yao Z, Keyes CC, et al. Genome-wide DNA methylation studies suggest distinct DNA methylation patterns in pediatric embryonal and alveolar rhabdomyosarcomas. Epigenetics 2012;7:400-8. [Crossref] [PubMed]


