
Critical evaluation of the subcutaneous engraftments of hormone naïve primary prostate cancer
Introduction
Patient-derived xenografts (PDXs) have become a preferred platform for preclinical drug testing. Successful PDXs from prostate cancer represent the parental tumor phenotype, genotype, and heterogeneity and enable in vivo interactions between tumor cells and relevant accompanying stroma (1,2). Cultured cancer cell lines are adapted to grow in vitro and often poorly predict human responses in clinical trials (3,4). For example, the three most commonly used prostate cancer cell lines (PC-3, DU-145, and LNCaP) do not express wild-type androgen receptor (AR) (5).
Serially transplantable human prostate cancer PDX lines have been created mostly from metastatic sites (6). They are grown either subcutaneously or under the subrenal capsule in immunodeficient mice (6). Orthotopic implantation in the prostate has also been reported to enable prostate cancer engraftments in mice (7). However, PDX models for non-metastatic prostate cancer are scarce. A recent consensus report summarized the available serially transplantable human prostate cancer xenograft lines, yet most of these originated from metastatic sites or represented advanced disease (6). Transplantable subcutaneously grown primary prostate cancer models from early androgen naïve disease are scarce, such as the PC-135 and PC-310 lines grown in nude mice with the support of testosterone (Te) pellets (6,8), and the two MD Anderson lines grown in intact SCID mice (6). Subrenal engraftment of primary hormone naïve prostate cancer has been more successful, resulting in nine Living Tumor Laboratory lines grown in NOD/SCID mice supplemented with Te (6,9). Most other prostate carcinoma PDX lines reviewed in the literature are from castration-resistant prostate cancer (CRPC) and therefore more clinically advanced. Only a few primary prostate cancer PDX models are available in international PDX repositories (i.e., Jackson Laboratories, NCI, Charles River, Champion, EuroPDX and PDXfinder).
Development of representative PDX models for non-metastatic prostate cancer is essential for studying the earliest disease events, as up to one-third of patients with local disease will eventually relapse. We have previously focused on subcutaneous (sc) (10-14), intratibial (15,16) and orthotopic (17-21) xenograft models of advanced prostate cancer. In the present study, we used 26 clinical primary prostate tumor specimens in 84 nude mice with the aim of producing a research model for hormone naïve primary prostate cancer. Of the grafting methodologies available to us, we chose to use the robust subcutaneous grafting technique, which can easily be transferred to other laboratories. Our results show that the first passage tumor grafts closely mimicked the genetic and immunohistochemical phenotypes and Gleason grades of their parental tumors. Expression levels of AR, prostate-specific antigen (PSA), and keratins were retained in engraftments, and a detailed genomic analysis revealed high fidelity of the engraftments with their corresponding primary tumors. With regard to our fairly large patient and mouse number, engraftments from only seven patients were able to retain primary carcinoma in the mice after first passage, while further passages resulted in rapid deterioration of the prostate tumor architectures. Histological validation of the exact area to be engrafted was imperative for the engraftment success. We could not show effect of recipient mouse immunocompromization level or patient characteristics on tumor engraftment rate. To our surprise, Te supplementation of the recipient mice correlated with a lower engraftment rate. Because several of our study patients eventually relapsed, we critically evaluated our and other existing methodologies and provide ideas for enhancing PDX creation from early stage prostate carcinoma.
Methods
Patient-derived tumor tissues
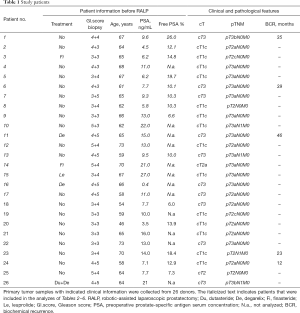
Clinical prostate tumor specimens were collected from robotic-assisted laparoscopic radical prostatectomies at Turku University Hospital (Turku, Finland) between April 2013 and November 2017 within Turku Prostate Cancer Consortium biobanking (Table 1). For biobanking purposes, an experienced uro-pathologist chose the presumed cancerous tissue area. Three samples (cylinders 0.5 or 0.8 cm in diameter) were drilled. Each cylinder was divided longitudinally into nine pieces and the location of the piece that was designed for implantation was recorded. The tissue was transferred immediately to the animal laboratory and implanted into mice. Later, H&E-stained validation sections adjacent to the engrafted samples were analyzed histologically for the percentage of carcinoma, benign epithelium, and stroma.
Full table
Tumor implantation
Six-week-old immunodeficient male mice (BALB/c nude, n=58, CAnN.Cg-Foxn1nu/Crl, Charles River, Germany; NOG mice, n=7, NOD.Cg-Prkdcscid Il2rgtm1Sug/JicTac Taconic, Denmark; NOD.SCID mice, n=6, NOD/MrkBomTac-Prkdc SCID, Taconic, Denmark; or SHrN hairless NOD.SCID, n=13, NOD.Cg-PrkdcscidHrhr/NCrHsd, Envigo, France, Table 7) were maintained under controlled pathogen-free conditions (20–21 °C, 30–60% relative humidity, 12-hour light cycle, soya-free chow, and autoclaved drinking water).
Full table
Full table
Full table

Full table
Full table
Full table
Tissue specimens were aseptically cut into 1–2 mm3 pieces in a laminar hood using razor blades. Tumor pieces were implanted subcutaneously into the inter-scapular fat pads of immunodeficient mice. A total of 33 mice were supplemented with 20–25 mg Te propionate pellets, which were pressed in-house from Te-propionate powder (Sigma-Aldrich). Nineteen mice were supplemented with 12.5 mg 5α-DHT pellets (Innovative Research of America, FL, USA). Thirty-two mice received no hormone pellets.
At sacrifice, gross necropsy was performed, and the findings were documented. Tumors were removed and collected for histology or transplanted further to second or third generation of mice using the same protocol as described above.
Histology and immunohistochemistry
Tissues were fixed with 4% neutral-buffered formalin. Samples were embedded in paraffin and 4-µm sections cut and stained with hematoxylin and eosin or used for immunohistochemical analyses.
For immunohistochemistry, sections were deparaffinized in xylene and rehydrated in a descending series of ethanol (100–70%). Endogenous peroxidase activity was blocked with 3% hydrogen peroxidase. Sections were incubated in humid chambers with primary antibodies detecting AR (Santa Cruz, SC-816, N-20, rabbit polyclonal IgG, 1:100), PSA (Novus Biologicals, NB110-59960, rabbit polyclonal IgG, 1:100), and pan-cytokeratin (ThermoScientific, MS-343-P, mouse monoclonal IgG, 1:100) overnight at 4 °C. Secondary antibodies were biotin-labeled goat anti-rabbit (Vector, Burlingame, USA), K405 Envision dual link system-HRP (DAKO), or horse anti-mouse (Vector, Burlingame, USA). Immunoperoxidase staining was performed using an ABC kit (Vector Laboratories, CA, USA), detected with diaminobenzidine (DAB), and eventually counterstained with Mayer’s hematoxylin.
Positive controls for each antibody and negative controls without primary antibody were used to verify the specificity of staining. Tissue morphology was evaluated by a trained pathologist (P Taimen). AR, PSA and pan-cytokeratin staining intensities were evaluated separately for cancer and healthy epithelium by three independent observers (M Valta, J Ylä-Pelto, J Tuomela). Results were reanalyzed until consensus was reached. Proportion of cancer, epithelium and stroma was calculated from H&E stained samples.
Blood samples and PSA measurement
Blood samples (200 µL) were collected from the saphenous vein after a warming procedure before implantation of tumor pieces, at sacrifice, and once in between. The serum samples were prepared within 1 hour of sampling and stored at −80 °C for PSA analysis (Quantikine Human Kallikrein 3/PSA ELISA kit, Minneapolis, R&D Systems). Serum PSA concentration was determined using the VICTOR2 Multilabel Counter (PerkinElmer, Waltham, MA, USA).
DNA isolation
DNA was isolated from tumors using the DNeasy Blood & Tissue kit (Qiagen GmbH, Hilden, Germany, Cat No./ID: 69506). Engrafted tumor DNA was isolated from paraffin-embedded samples. Before isolation, paraffin was removed using xylene. After paraffin removal, all samples were handled similarly. Briefly, tissues were cut into small pieces and incubated in lysis buffer overnight at 56 °C before following the DNA isolation protocol described by the manufacturer. The quality of the genomic DNA was confirmed using an Advanced Analytical Fragment Analyzer. Sample concentrations were measured using Nanodrop ND-2000 (Thermo Scientific) and Qubit® Fluorometric Quantitation (Life Technologies). DNA samples had genomic quality numbers (GQNs) of 4.0–8.2. We selected five primary tumors and seven of their corresponding engrafted tumors for sequencing. The selection was based on DNA quality and availability.
NGS library preparation and sequencing
Library preparation was performed using the TruSeq Exome Library Preparation kit (Illumina) according to the manufacturer’s instructions. Library quality was confirmed using Fragment Analyzer (Advanced Analytical) and the concentrations quantified using Qubit® Fluorometric Quantitation (Life Technologies). Libraries were sequenced in a HiSeq 3000 instrument (Illumina) using paired-end sequencing and a 75 bp read length. Library preparation and sequencing were performed at the Finnish Functional Genomics Centre (FFGC), University of Turku, Åbo Akademi University, and Biocenter Finland.
Data extraction
Base calling was performed using Illumina’s standard bcl2fastq2 software with automatic adapter trimming.
Exome sequencing data analysis and variant calling
The quality of the raw exome sequencing reads was assessed using FASTQC (22) with all samples passing the initial quality control. In order to examine the portion of mouse gDNA in the tumor samples, we first aligned the reads to a combined reference consisting of concatenated mouse (mm10) and human (Hg19) reference sequences using BWA-mem (23) and a previously described technique (24). The fraction of mouse gDNA in the tumor samples varied from 0.52% to 8.98%. All reads that aligned to the mouse reference genome were removed from the samples. We followed the Genome Analysis Toolkit (GATK) Best Practices for all proceeding steps, including sequence realignment and duplicate detection (25). The resulting alignment files were used to detect copy number variant (CNV) regions with Control-FREEC (26).
Statistical analysis
The Pearson correlation for CNVs was calculated for all tumor pairs. For single nucleotide polymorphism (SNP) and small indel calling, GATK HaplotypeCaller was used, followed by VariantFiltration to filter variants as a final step to achieve the final sample-wise variant lists. The additional threshold for a read depth ≥8 at variant sites was used to increase the reliability of the variant calling of the FFPE samples.
The effects of Gleason score, cT, pTNM, biochemical relapse, age at diagnosis, PSA, carcinoma in adjacent prostate tissue, mouse strain, hormone pellet, engraftment time, on tumor take were analyzed using Chi-square test in Graph Pad Prism V6.0. Tissue composition between primary tumor and engraftment, were studied using unpaired t-test in Graph Pad Prism V6.0.
Results
Most carcinomas developed palpable engraftments
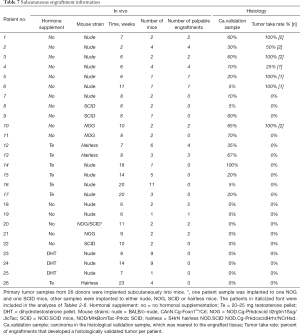
Tissue samples were collected from 26 donors who underwent radical prostatectomy (Table 1). All samples were collected from primary non-metastatic prostate cancer and 20 out of 26 patients were hormone naïve. The tissues were implanted subcutaneously into a total number of 84 mice (one patient sample to 1–11 mice according to tissue availability, Table 7). Engraftments were removed 2–20 weeks (mean 10 weeks) after implantation.
Thirteen patient samples developed a palpable engraftment (Table 7). Palpable engraftments from seven patients formed histologically confirmed adenocarcinomas in 11 mice. The remaining palpable engraftments were composed of only benign prostatic epithelium and stroma.
Presence of carcinoma in adjacent histological validation samples was imperative for tumor formation
H&E-stained primary tumor validation sections adjacent to the implanted samples were analyzed histologically by a pathologist after engraftment, as soon as they were available. Out of 26 patient samples, 17 had carcinoma in their validation sample (Table 7). Prostate cancer samples engrafted as histological tumors only if the adjacent validation section in the primary tumor site contained cancerous tissue (5–100%), and this correlation was statistically significant (P<0.01). By contrast, if the validation sections adjacent to the engraftment sample site were devoid of carcinoma, the engraftments contained only viable normal prostatic tissue. However, the presence of carcinoma in the validation sample did not always induce tumor growth in the engraftment, as seen with ten patient samples.
We defined tumor take rate as the percentage of patients, who had carcinoma in the validation sample (n=17), and who developed at least one histologically validated tumor (n=7). Thus, the tumor take rate in this study was 41%. Six out of these seven patients were hormone naïve. Patients with no cancer in the validation sample were excluded from further analyses.
Engraftments share the characteristics of their parental tumors
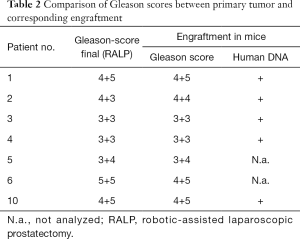
Next, the pathological and morphological features of the primary prostate tumors were compared to the corresponding first-passage engraftments. Morphologically, engraftments and primary tumors closely resembled each other (Figure 1, H&E staining) and the Gleason scores were nearly identical. DNA sequencing further showed that all the analyzed samples contained human DNA (Table 2).
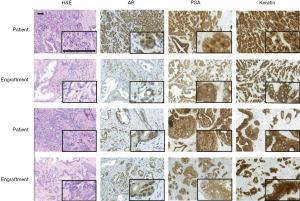
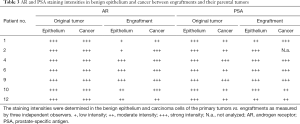
Immunohistochemistry was used to study AR, PSA, and pan-cytokeratin protein expression in the primary tumors and their corresponding engraftments (Figure 1, Table 3). In engraftments originating from patients 1 and 12, AR expression decreased to some extent in cancer cells but was still clearly detectable. On the other hand, PSA expression in engraftments from patients 1 and 6 was higher compared with the original tumor specimens. In all the other engraftments, AR and PSA expression was conserved. Keratin (4, 5, 6, 7, 8, 10, 13, 14, 18, and 19) expression was retained in all cancerous and non-cancerous samples. Serum PSA concentration was below the limit of detection in all the mice (data not shown), suggesting that the number of engrafted PSA-secreting epithelial or tumor cells was too small to induce a systemic elevation of serum PSA.
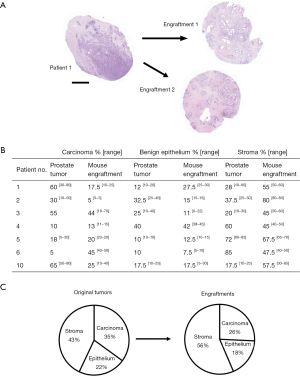
Then we compared the changes in histological composition between the primary tumors and engraftments in more detail. For this purpose, the validation samples (at sites adjacent to engraftment) were analyzed microscopically with regard to the relative percentages of carcinoma, benign epithelium, and stroma (Figure 2A). We found that the percentage of carcinoma decreased in four cases and increased in three cases relative to their parental tumor specimens (Figure 2B). On average, engraftments contained 26% less cancer than their primary tumor counterparts. The percentage of normal epithelium decreased in three cases, increased in three cases and remained similar in one case compared with the original tumor. Engraftments contained on average 18% less normal epithelium than their respective primary tumors. The percentage of stroma decreased in two cases and increased in five cases. Altogether, we found a statistically significant 33% increase in stromal content compared to primary tumors (P<0.05). These results indicate good preservation of all three main histological compartments (benign epithelium, stroma, and carcinoma) in the engraftments, but a modest increase in the proportion of stromal components at the expense of carcinoma occurred during the follow-up period (Figure 2C).
Genomic assessment of engraftments and corresponding primary tumors
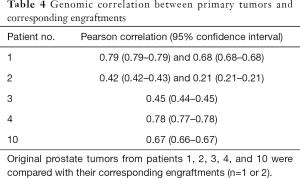
Next, we analyzed the preservation of the molecular composition of the engraftments compared to their parental tumors. When DNA exome sequencing data were analyzed, mouse and human DNA were separated so that the overlapping areas between Homo sapiens and Mus musculus sequences were removed from the analysis. In general, the CNVs between the engraftments and corresponding tumor samples had a relatively high correlation (Pearson correlation coefficient up to 0.79 with P values <1.0×10−5 and on average 0.57, Table 4). In addition, we assessed the differences in SNPs and small indels among 19 cancer-associated genes (Table 5), finding 44 variants shared by each tumor pair. None of the variants differed consistently between the engraftments and corresponding primary tumor samples.
Mouse hormone supplementation or immunodeficiency do not promote the histologically detectable tumors
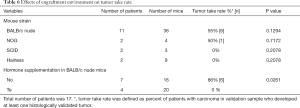
We engrafted tumors from 11 patients into 36 BALB/c nude mice and tumors from six patients into 16 mice that were more immunocompromised compared to nude mice (NOG, SCID or hairless mouse strains) (Table 6). Six patient samples produced histologically detectable tumors in BALB/c nude mice, whereas only one patient sample produced a histologically confirmed tumor in the more immunocompromised mice. Although this suggests that a more severely immunodeficient background (BALB/c nude vs. NOG, SCID or hairless mice) does not favor the preservation of adenocarcinomas, these differences failed to reach statistical significance but due to a small number of samples.
Samples from seven patients were engrafted into 16 nude mice without hormone supplementation, samples from four patients to 20 Te pellets-bearing nude mice (Table 6). In addition, samples from three patients were implanted to 19 nude mice bearing DHT pellets (Table 6) but they were excluded from analysis due to lack of adenocarcinoma in validation sample. Surprisingly, 6 out of 7 patient samples in mice without hormone supplementation produced adenocarcinomas, whereas the success rate of Te supplementation was 0% (Table 6). This difference between the tumor-bearing nude mice with or without hormone supplementation was statistically significant (P=0.0261). Samples from four patients were engrafted into seven more immunocompromised mice (NOG, SCID or hairless) without hormone supplementation, and samples from two patients into nine with Te supplementation. In this comparison, only one patient sample produced adenocarcinoma in mice (i.e., NOG mice) without hormone supplementation while no adenocarcinomas were observed in Te-supplementation group.
In general, engraftment time, Gleason score, clinical stage, pathological stage, patient age, patient treatment or preoperative PSA had no statistically significant effect on the tumor take rate (results not shown). Interestingly, tumor samples from 2/3 patients with biochemical relapse formed successful engraftments.
The viability of adenocarcinoma in engraftments may be limited to first generation tumors
Engraftments originating from five patients were also subjected to a second passage of subcutaneous implantation (n=19 mice), and two of them to a third round (n=3 mice). Microscopic analyses revealed small cancerous areas in a few of the second-generation tumors, whereas the third-generation engraftments did not contain any cancer cells (results not shown). Instead, the second and third round engraftments typically exhibited normal or metaplastic benign prostate glands with frequent degenerative changes, such as cystic dilatation, fibrosis, and increased stromal elements.
Discussion
A small number of cell lines has been established from prostate cancer tissues, and their application to preclinical studies has been limited due to their characteristics that usually differ from their original tumors. Recent technological improvements have increased the success rates for the establishment of such PDX models, which retain the characteristics of the original tumors and tumor heterogeneity. However, most of these models, for example the well-characterized and representative LuCap xenograft lines (2), originate mainly from metastatic prostate cancer tissues. Non-metastatic primary hormone naïve prostate carcinoma is not well represented, yet many such patients will eventually relapse. We implanted prostate cancer tissue from mostly hormone naïve 26 patients subcutaneously to 84 immunodeficient mice. Subcutaneous implantation method was chosen, because we wanted to evaluate a predictive platform for drug response studies that could be easily transferred to other laboratories. Both the genotypes and phenotypes of the engraftments closely resembled their parental tumors, indicating that these engraftments retained the properties of the primary tumors after implantation.
Histologically detectable cancer in engraftments resembled their parental tumors with regard to immunohistochemical expression of AR, PSA, and keratin. Furthermore, Gleason grades between the primary tumors and their corresponding engraftments were nearly identical, similar to studies with orthotopic tumors (7). We showed that all three main histological compartments (benign epithelium, stroma, and carcinoma) were well preserved in the engraftments but the relative proportion of stroma was modestly increased in engraftments compared to corresponding primary tumors, matching other findings (27).
Little is known about genetic changes that occur during PDX propagation, whereas in vitro cultured cell lines are known to differ from their in vivo counterparts. Previous studies have shown that human tumors engrafted in mouse hosts retain therapeutically relevant genomic aberrations found in the original patient tumors (28). There are also reports suggesting that serially transplantable PDXs are genomically unstable and transform during the first few in vivo passages (29,30). However, a genome-wide analysis of CNVs between our engraftments and corresponding primary tumor samples revealed a relatively high correlation (Table 4). Moreover, we found no consistent differences in the variation of exome regions of 19 cancer-associated genes between the engraftments and their parental tumors. Thus, our results do not support the hypothesis of rapid genetic changes during engraftment.
Not surprisingly, the presence of carcinoma in the validation tissue was imperative for carcinoma preservation of engraftments, which highlights the importance for carefully analysing the exact primary tumor area to be implanted. Similarly, it was previously noticed that CRPC transurethral resection of the prostate (TURP) tissue engrafted successfully under the subrenal capsule in mice, only if a high proportion of viable cancer cells was present in the original specimen (31).
The generation of serially transplantable prostate cancer xenograft lines from primary hormone naïve specimens has rarely been reported with the subcutaneous model. In a previous study, less than 2% of the immunodeficient BALB/c nude mice developed a PDX, when subcutaneous implantation of fresh prostate cancer specimens was used (8). In another study, Priolo et al. (27) observed subcutaneous engraftment of primary prostate tumors in only 1 out of 6 nude mice implanted with Te pellets. In a recent study of 147 primary prostate cancer patient samples grown subcutaneously in severely immunocompromised Rag2−/−γC−/− mice with Te supplementation, only three of the engrafted biopsies resulted in a stable line, and all these three were derived from CRPC or hormonally treated patients (32). We observed 41% tumor take rate in the first passage, but the preservation of cancerous tissue in the engraftments could not be expanded beyond further passages. Improved diagnostics has resulted in primary prostate cancers often being excised at an early stage, when the tumor material is scarce (33). In addition, a proportion of prostate cancer patients have a slow-growing cancer, which could have been managed through active surveillance. A minuscule percentage of carcinoma in the original material combined with this latent and heterogenous nature of prostate tumors greatly diminishes the possibility of PDX engraftment success. Because of the diminishing sizes of prostate tumors, a more careful analysis of the starting material is necessary for successful engraftment compared to earlier studies. The use of frozen sections of the implanted tissue or MRI images of the primary prostate tumor could aid the success of hitting the most aggressive cancerous lesions and therefore increase the likelihood of adenocarcinoma preservation in the engraftments also subcutaneously.
An attractive alternative for successful primary prostate cancer PDX generation is the use of more immunodeficient mouse strains. BALB/c nude mice still retain humoral adaptive immune response to some degree and an intact innate immune system (34), and tumor regression caused by the remaining immune surveillance may reduce engraftment of PDXs. The maturation of B and T cells is more severely impaired in SCID or NOD/SCID mice compared to nude mice, and NOD/SCID mice also harbor a defect in the innate immune system (35). In addition to these traits, the so-called triple immunodeficient NSG (NOD SCID gamma) and SHrN hairless NOD.SCID mice have reduced cytokine signaling (36). Though higher PDX engraftment rates have been observed for prostate cancer in some studies using these more immunodeficient strains (6), we and others did not find benefit from using them (Tables 6,7) (27). The outgrowth of Epstein-Barr virus (EBV)-transformed human lymphomas in highly immunocompromised mouse strains observed by us (unpublished results) and others (32,37) is a severe limitation for their use. In addition, immunodeficient background practically prevents immunotherapy studies, which have recently gained attention for prostate cancer (38). It would be intriguing to use a more sophisticated background, such as immunologically humanized mice for our studies. Humanized mice are established by grafting tumor tissue into mice engrafted with human immune cells (39), which allows investigation of the role of the human immune system in PDX growth, as well as the efficacy of immune-based therapies. However, the applicability of this model is currently limited by its high cost.
The successful MD Anderson hormone naïve primary prostate cancer xenografts were inoculated subcutaneously into SCID mice without additional Te supplementation (6). We used BALB/c nude, NOG, SCID and hairless mice that were either intact or supplemented with hormone pellets to determine, whether this would increase tumor engraftment. We chose BALB/c nude and hairless mice, because tumor growth is easier to follow in them than in fur-wearing NOG and NOD.SCID mice. Surprisingly, when tumor tissue was implanted into BALB/c nude mice, histological adenocarcinoma was found in nine out of 16 mice without hormone supplementation, but in none of the 20 mice with hormone pellet. Similar tendency was observed in the more immunocompromised mice. Nude mice are known to have low and highly variable circulating levels of serum Te comparable to elderly men (8,40), and the majority of our patients were hormone naïve and elderly. Our results are in line with earlier xenograft studies (41), indicating that high serum androgen levels do not necessarily promote tumor growth in animals. It is possible that for the generation of primary hormone naïve prostate cancer xenografts in nude mice, no supplementary Te is needed.
Obviously, tumor microenvironment plays an important role in tumor progression and requires more attention. Subrenal implantation of prostate cancer tissues has been successful for the generation of transplantable PDXs even from lower grade primary prostate cancer, presumably due to the rich vasculature and supply of nutrients (9). This method requires significantly more technological experience, resources, and time, and is not effectively transferred between laboratories. For example, an average 22-month median initial latency period was necessary for the Living Tumor Laboratory PDX lines from primary low-grade prostate cancer (9). The use of an orthotopic model for creating engraftments of primary prostate tumors has yielded a high engraftment rate of 55.6%, though this model was not studied for its transplantability (7). In further studies, the human tumor microenvironment could also be mimicked by inoculating the digested primary tumor cell suspension into Myogel, which is a total protein extract formulated from human myoma (42). Toivanen et al. (43) developed a protocol in which co-grafting of early-stage primary prostate tumors with mouse neonatal stroma results in increased engraftment rate and growth in vivo already within the 8-week study protocol. An interesting option for a bone metastases model is co-grafting tumor cells on viable human subchondral bone discs into NOD.SCID mice (44).
There is a clear unmet need for representative prognostic and predictive preclinical models of primary low grade, hormone naïve prostate cancer. Despite various attempts, only subrenal implantation of primary prostate cancer has yielded serially transplantable PDX models, with a grafting period of almost 2 years. In our model, patient tissue engraftments retained both the genotypes and phenotypes, as well as Gleason grades of their parental tumors during the 2-month study period. Therefore, our subcutaneous engraftments of primary hormone naïve prostate cancer have a great potential for a representative personalized model of primary prostate cancer. The application of our model for personalized medicine will still require better pre-evaluation of engrafted tissue, which will be achieved by tight collaboration between urologists, pathologists and basic scientists.
Acknowledgments
The authors warmly thank Mr. Jaakko Lehtimäki and the personnel of Auria Biobank and Pharmatest Services Ltd. for excellent technical assistance.
English of the manuscript has been edited by San Francisco Edit.
Funding: This study was funded by the K. Albin Johansson Foundation (J Tuomela), Cancer Organization of South West Finland (J Tuomela), Maud Kuistila Memorial Foundation (J Tuomela), the Academy of Finland (grant 287040 to PJ Koskinen), and State Research Funding to the responsibility area of Turku University Hospital (P Härkönen and M Valta). P Taimen was supported by the Academy of Finland Clinical Researcher funding and the Finnish Cancer Society. This study was also supported by Finnish Functional Genomics Centre, University of Turku and Åbo Akademi, ELIXIR Finland and Biocenter Finland.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tau.2020.03.38). PT reports grants from Academy of Finland, grants from Finnish Cancer Society, during the conduct of the study; personal fees from Roche, outside the submitted work. PJK reports grants from Academy of Finland, during the conduct of the study. PH reports grants from State Research Funding to the responsibility area of Turku University Hospital, during the conduct of the study. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was approved by the ethical committee of the University of Turku and the Scientific Advisory Board of Auria Biobank (the registration numbers: ETMK: 3/180/2013, PH806A) and informed consent was taken from all the patients. Animals were cared for in accordance with the Directive 2010/63/EU of the European Parliament and the Council for the Care and Use of Laboratory Animals (license numbers for animal experiments: ESAVI6057/04.10.03/2011, ESAVI3257/04.10.07/2014, and ESAVI2329/04.10.07/2017).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Davies AH, Wang Y, Zoubeidi A. Patient-derived xenografts: A platform for accelerating translational research in prostate cancer. Mol Cell Endocrinol 2018;462:17-24. [Crossref] [PubMed]
- Nguyen HM, Vessella RL, Morrissey C, et al. LuCaP Prostate Cancer Patient-Derived Xenografts Reflect the Molecular Heterogeneity of Advanced Disease an--d Serve as Models for Evaluating Cancer Therapeutics. Prostate 2017;77:654-71. [Crossref] [PubMed]
- Choi SY, Lin D, Gout PW, et al. Lessons from patient-derived xenografts for better in vitro modeling of human cancer. Adv Drug Deliv Rev 2014;79-80:222-37. [Crossref] [PubMed]
- Ocana A, Pandiella A, Siu LL, et al. Preclinical development of molecular-targeted agents for cancer. Nat Rev Clin Oncol 2010;8:200-9. [Crossref] [PubMed]
- Sobel RE, Sadar MD. Cell lines used in prostate cancer research: a compendium of old and new lines--part 1. J Urol 2005;173:342-59. [Crossref] [PubMed]
- Navone NM, van Weerden WM, Vessella RL, et al. Movember GAP1 PDX project: An international collection of serially transplantable prostate cancer patient-derived xenograft (PDX) models. Prostate 2018;78:1262-82. [Crossref] [PubMed]
- Saar M, Körbel C, Linxweiler J, et al. Orthotopic tumorgrafts in nude mice: A new method to study human prostate cancer. Prostate 2015;75:1526-37. [Crossref] [PubMed]
- van Weerden WM, de Ridder CM, Verdaasdonk CL, et al. Development of seven new human prostate tumor xenograft models and their histopathological characterization. Am J Pathol 1996;149:1055-62. [PubMed]
- Lin D, Wyatt AW, Xue H, et al. High fidelity patient-derived xenografts for accelerating prostate cancer discovery and drug development. Cancer Res 2014;74:1272-83. [Crossref] [PubMed]
- Minn H, Grönroos TJ, Komar G, et al. Imaging of tumor hypoxia to predict treatment sensitivity. Curr Pharm Des 2008;14:2932-42. [Crossref] [PubMed]
- Wegiel B, Bjartell A, Tuomela J, et al. Multiple cellular mechanisms related to cyclin A1 in prostate cancer invasion and metastasis. J Natl Cancer Inst 2008;100:1022-36. [Crossref] [PubMed]
- Tuomela J, Grönroos TJ, Valta MP, et al. Fast growth associated with aberrant vasculature and hypoxia in fibroblast growth factor 8b (FGF8b) over-expressing PC-3 prostate tumour xenografts. BMC Cancer 2010;10:596. [Crossref] [PubMed]
- Yu L, Toriseva M, Tuomala M, et al. Increased expression of fibroblast growth factor 13 in prostate cancer is associated with shortened time to biochemical recurrence after radical prostatectomy. Int J Cancer 2016;139:140-52. [Crossref] [PubMed]
- Virtanen SS, Ishizu T, Sandholm JA, et al. Alendronate-induced disruption of actin cytoskeleton and inhibition of migration/invasion are associated with cofilin downregulation in PC-3 prostate cancer cells. Oncotarget 2018;9:32593-608. [Crossref] [PubMed]
- Valta MP, Tuomela J, Bjartell A, et al. FGF-8 is involved in bone metastasis of prostate cancer. Int J Cancer 2008;123:22-31. [Crossref] [PubMed]
- Valta MP, Zhao H, Saar M, et al. Spheroid culture of LuCaP 136 patient-derived xenograft enables versatile preclinical models of prostate cancer. Clin Exp Metastasis 2016;33:325-37. [Crossref] [PubMed]
- Tuomela JM, Valta MP, Väänänen K, et al. Alendronate decreases orthotopic PC-3 prostate tumor growth and metastasis to prostate-draining lymph nodes in nude mice. BMC Cancer 2008;8:81. [Crossref] [PubMed]
- Tuomela J, Valta M, Seppänen J, et al. Overexpression of vascular endothelial growth factor C increases growth and alters the metastatic pattern of orthotopic PC-3 prostate tumors. BMC Cancer 2009;9:362. [Crossref] [PubMed]
- Valta MP, Tuomela J, Vuorikoski H, et al. FGF-8b induces growth and rich vascularization in an orthotopic PC-3 model of prostate cancer. J Cell Biochem 2009;107:769-84. [Crossref] [PubMed]
- Santio NM, Eerola SK, Paatero I, et al. Pim Kinases Promote Migration and Metastatic Growth of Prostate Cancer Xenografts. PLoS One 2015;10:e0130340. [Crossref] [PubMed]
- Martikainen M, Ruotsalainen J, Tuomela J, et al. Oncolytic alphavirus SFV-VA7 efficiently eradicates subcutaneous and orthotopic human prostate tumours in mice. Br J Cancer 2017;117:51-5. [Crossref] [PubMed]
- Andrews S. FastQC: a quality control tool for high throughput sequence data. 2010. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc
- Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv:1303.3997v1 [q-bio.GN]. 2013.
- Callari M, Batra AS, Batra RN, et al. Computational approach to discriminate human and mouse sequences in patient-derived tumour xenografts. BMC Genomics 2018;19:19. [Crossref] [PubMed]
- Van der Auwera GA, Carneiro MO, et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics 2013;43:11.10.1-11.10.33.
- Boeva V, Popova T, Bleakley K, et al. Control-FREEC: a tool for assessing copy number and allelic content using next-generation sequencing data. Bioinformatics 2012;28:423-5. [Crossref] [PubMed]
- Priolo C, Agostini M, Vena N, et al. Establishment and genomic characterization of mouse xenografts of human primary prostate tumors. Am J Pathol 2010;176:1901-13. [Crossref] [PubMed]
- Woo XY, Srivastava A, Graber JH, et al. Genomic data analysis workflows for tumors from patient-derived xenografts (PDXs): challenges and guidelines. BMC Med Genomics 2019;12:92. [Crossref] [PubMed]
- Ben-David U, Ha G, Tseng YY, et al. Patient-derived xenografts undergo mouse-specific tumor evolution. Nat Genet 2017;49:1567-75. [Crossref] [PubMed]
- Eirew P, Steif A, Khattra J, et al. Dynamics of genomic clones in breast cancer patient xenografts at single-cell resolution. Nature 2015;518:422-6. [Crossref] [PubMed]
- Lawrence MG, Pook DW, Wang H, et al. Establishment of primary patient-derived xenografts of palliative TURP specimens to study castrate-resistant prostate cancer. Prostate 2015;75:1475-83. [Crossref] [PubMed]
- Taurozzi AJ, Beekharry R, Wantoch M, et al. Spontaneous development of Epstein-Barr Virus associated human lymphomas in a prostate cancer xenograft program. PLoS One 2017;12:e0188228. [Crossref] [PubMed]
- Stamey TA, Caldwell M, McNeal JE, et al. The prostate specific antigen era in the United States is over for prostate cancer: what happened in the last 20 years? J Urol 2004;172:1297-301. [Crossref] [PubMed]
- Hanna N, Davis TW, Fidler IJ. Environmental and genetic factors determine the level of NK activity of nude mice and affect their suitability as models for experimental metastasis. Int J Cancer 1982;30:371-6. [Crossref] [PubMed]
- Beck JA, Lloyd S, Hafezparast M, et al. Genealogies of mouse inbred strains. Nat Genet 2000;24:23-5. [Crossref] [PubMed]
- Shultz LD, Lyons BL, Burzenski LM, et al. Human lymphoid and myeloid cell development in NOD/LtSz-scid IL2R gamma null mice engrafted with mobilized human hemopoietic stem cells. J Immunol 2005;174:6477-89. [Crossref] [PubMed]
- Wetterauer C, Vlajnic T, Schüler J, et al. Early development of human lymphomas in a prostate cancer xenograft program using triple knock-out immunocompromised mice. Prostate 2015;75:585-92. [Crossref] [PubMed]
- Graff JN, Alumkal JJ, Drake CG, et al. Early evidence of anti-PD-1 activity in enzalutamide-resistant prostate cancer. Oncotarget 2016;7:52810-7. [Crossref] [PubMed]
- Shultz LD, Brehm MA, Garcia-Martinez JV, et al. Humanized mice for immune system investigation: progress, promise and challenges. Nat Rev Immunol 2012;12:786-98. [Crossref] [PubMed]
- Michiel Sedelaar JP, Dalrymple SS, et al. Of mice and men--warning: intact versus castrated adult male mice as xenograft hosts are equivalent to hypogonadal versus abiraterone treated aging human males, respectively. Prostate 2013;73:1316-25. [Crossref] [PubMed]
- Song W, Soni V, Soni S, et al. Testosterone inhibits the growth of prostate cancer xenografts in nude mice. BMC Cancer 2017;17:635. [Crossref] [PubMed]
- Salo T, Sutinen M, Hoque Apu E, et al. A novel human leiomyoma tissue derived matrix for cell culture studies. BMC Cancer 2015;15:981. [Crossref] [PubMed]
- Toivanen R, Frydenberg M, Murphy D, et al. A preclinical xenograft model identifies castration-tolerant cancer-repopulating cells in localized prostate tumors. Sci Transl Med 2013;5:187ra71. [Crossref] [PubMed]
- Holen I, Nutter F, Wilkinson JM, et al. Human breast cancer bone metastasis in vitro and in vivo: a novel 3D model system for studies of tumour cell-bone cell interactions. Clin Exp Metastasis 2015;32:689-702. [Crossref] [PubMed]


