
Defining new genetic etiologies of male infertility: progress and future prospects
Introduction
Approximately 15% of couples trying to conceive are infertile, and male factors are responsible for 40–50% of these cases (1,2). The diagnosis of male fertility is usually performed through the observation of sperm number, motility, and morphology under the microscope (2). Nevertheless, the clinical value of semen analysis (or seminogram) has been called into question (3,4). Therefore, the identification of new, predictive and effective male infertility markers is a must, and several multicenter projects and international consortia are currently actively searching for new male infertility markers in the context of genetics and epigenetics.
The aim of this review is to highlight the most important advances in the field of genetics of male infertility, to describe the main methodologies and techniques for identification and validation of novel infertility markers, to review current efforts being made to improve the diagnosis and treatment of male infertility, and to discuss how recent and future findings will advance clinical practice in the near future.
Established genetic components of male infertility
Disease-associated genetic variants can be broadly categorized as highly penetrant monogenic and associated variants. Highly penetrant monogenic variants are those which disrupt function of a single gene and result in a consistent phenotype, while associated variants may be present in affected or unaffected individuals, but the variant frequency is significantly associated with the disease state, and the variant may not directly impact gene function.
In the context of male infertility, several genetic causes of spermatogenic impairment are well established and already in clinical use, while others have been described more recently and vary in current clinical evidence (Table 1). The vast majority of assisted reproductive technology (ART) centers and reproductive urologists routinely utilize genetic screening in the diagnostic workup of infertile men, particularly those displaying azoospermia or severe oligozoospermia.
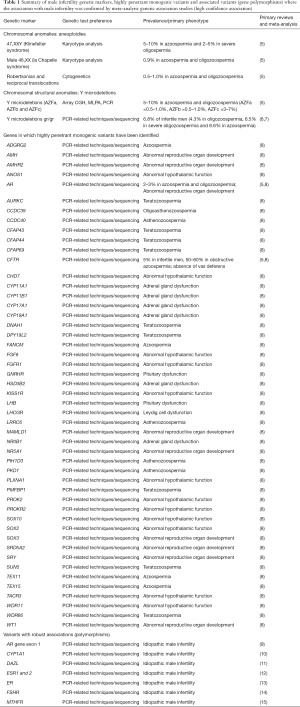
Full table
Highly penetrant monogenic variants
For example, established guidelines (16) indicate that infertile patients with non-obstructive azoospermia (NOA) and/or severe oligozoospermia should undergo cytogenetic testing (karyotype) and Y chromosome deletion testing due to the high prevalence of Klinefelter (47,XXY) syndrome [5–15% of NOA and severe oligozoospermic men (17)] and Y chromosome microdeletions or AZF deletions [5–10% of NOA and severe oligozoospermic men; 0.5% of infertile patients (17)]. Less frequent causes of male infertility include 46,XX male (1:20,000 new born; 0.9% of NOA men) or the Robertsonian translocations, inversions, and reciprocal translocations, ten-fold more frequent in men with oligozoospermia than in men with normozoospermia (5,6).
In the context of obstructive azoospermia, CFTR mutations are the main cause for agenesis of the vas deferens or obstruction. Subjects with CFTR mutations are excellent candidates for ICSI, using sperm retrieved from testis or epididymis because spermatogenesis in these patients is normal. The prevalence of CFTR mutations is about 5% in infertile patients and 50–60% in obstructive azoospermic patients (5).
In addition to Klinefelter’s syndrome, AZF deletions and CFTR mutations, only a small number of gene variants have reached the level of high clinical validity in the field of Andrology, however that number is growing as genome-wide approaches are applied increasingly in male infertility research.
Several genes that harbor variants with very strong evidence for a highly penetrant monogenic role in male infertility include ANOS1, AR, CFTR, CHD7, CYP11B1, FANCM, SRY, STAG2, and TEX11, in NOA; DPY19L2 and SPATA16 associated with globozoospermia; AMH, AMHR2, CYP11A1, CYP17A1, CYP19A1, CYP21A2, FGFR1, GNRHR, KISS1R, LHB, LHCGR, NR0B1, NR5A1, PROKR2, SRD5A2, TACR3, and WT1 associated with low or very low sperm count; and AURKC, DNAH1, CFAP43 and CFAP44, SUN5, WDR66 associated with multiple morphological abnormalities of the flagellum (MMAF) (8).
In current clinical practice, the screening for AR (androgen receptor) mutations has been introduced in some selected instances of quantitative sperm disorders due to the prevalence in infertility patients. Androgens and androgen receptors are essential for normal male sexual development before birth and during puberty and are crucial for the maintenance of the male phenotype and spermatogenesis. A mutation in AR [situated on the X chromosome (Xq11-12)], can cause androgen insensitivity syndrome. More than 1,000 mutations in AR gene have been described and the prevalence of clinically relevant AR mutations in azoospermic and oligozoospermic men is about 2–3% (5).
The list of highly penetrant monogenic variants with strong evidence for a role in male infertility has grown rapidly in the past few years and will continue to grow at an accelerated pace, largely driven by whole genome and exome sequencing approaches coupled with larger sample sets and carefully selected familial cases of infertility (6,8).
Associated variants
Moreover, some gene polymorphisms are considered potential risk factors for spermatogenic failure, or they may display association with male infertility without directly impacting gene function. The list of well validated variants includes variants in exon 1 of AR (9), CYP1A1 (10), DAZL (11), ESR1 and 2 (12), ER (13), FSHR (14), and MTHFR (15) polymorphisms among others, due to the existence of meta-analysis certifying the positive associations with male infertility (5,18). Gr/gr deletions likewise have been shown to be significantly associated with male infertility based on several large studies and meta-analyses. Smaller than AZF deletions, gr/gr deletions remove only a small part of the AZFc region. The prevalence of these microdeletions is estimated to be 6.8% in infertile men (4.3% in oligozoospermic men, 6.5% in men with severe oligozoospermia and 8.6% in azoospermic men) and 3.9% in controls (7), though frequencies vary by ethnicity and Y haplogroup (19).
The efficiency with which new disease-highly penetrant monogenic or associated variants will be discovered in the context of male infertility largely depends upon a handful of factors. These factors include the use of genome-wide approaches that enable the discovery of rare variants, careful phenotyping for precise classification of infertility phenotypes and large-scale collaborative studies to leverage the cumulative resources (patient samples along with assay and analytical resources) of multiple groups with a common goal.
Tools for genomic discovery
There is a long and growing list of genomic tools available for the discovery of novel genetic variants associated with male infertility. These include PCR- and array-based approaches as well as more recently refined whole exome and whole genome sequencing (WGS) approaches.
Conventional techniques
CGH arrays
The comparative genomic hybridization array (aCGH), is a well-stablished molecular cytogenetic method for the analysis of copy number variations (CNVs), which include submicroscopic insertions and deletions in the genome that can disrupt gene function. aCGH is used in some ART clinics to detect aneuploidies, well-characterized microdeletion/microduplication syndromes and sub-telomeric or other unbalanced chromosomal rearrangements (20). However, this technique is not able to identify balanced chromosomal alterations such as translocations and inversions. Using aCGH, the risk variant TEX11 was described only 5 years ago as cause of meiotic arrest and azoospermia in infertile men (21).
In aCGH, genomic DNA of the patient and control are differentially labeled with fluorescent probes such as Cyanine 3 (Cy3) and Cy5, following which equal amounts of labeled genomic DNA from a test and a reference sample are co-hybridized to an array containing the DNA targets, and the slides are scanned into image files using a microarray scanner. The spot intensities are measured, and the ratio of the fluorescence intensities is proportional to the ratio of the numbers of copies of specific DNA sequences. If there is an altered Cy3:Cy5 ratio, this indicates a loss or a gain of patient DNA at that specific genomic region.
MLPA technique
Genomic imbalances detected by aCGH are usually validated with other cytogenetic and molecular methods that can include customized multiplex ligation-dependent probe amplification (MLPA) assays (22). These analyses are based on multiplexed size-separation of the amplification products of a maximum of 40–50 target sequences in parallel in a single PCR reaction. Nowadays, more than 300 probe sets are commercially available and are specific for a very large range of common and rare genetic disorders. The main advantage of MLPA is the low cost of the technique (23). In the case of MLPA technique, one study allowed the description of some mutations in DMRT1 gene as the associate variant (24).
SNP arrays
Single nucleotide polymorphism (SNP) arrays are routinely used to detect common polymorphisms within a population. SNP arrays enable the assessment of hundreds of thousands to millions of polymorphisms across the genome to determine whether specific SNPs occur more frequently in a disease cohort compared with controls. They are most frequently used for genome-wide association studies of common polymorphisms but can also be used to detect CNVs based on probe intensities. In the past decade SNP arrays have been relatively widely employed in the study of male infertility, which has been important in characterizing the genetic architecture of the disease (see Table 1).
Next generation sequencing approaches
With significant advances in next generation sequencing (NGS) technologies and rapidly increased adoption of NGS, sequencing costs have decreased rapidly. This has resulted in significant shifts in the tools employed for disease variant discovery (25). While there is still some utility in characterizing frequencies of common polymorphisms and CNVs, WGS and whole exome sequencing (WES) have largely supplanted array-based technologies for gene discovery. WGS, as the name suggests, attempts to sequence the entire genome, however, technically the procedures cover about 95–96% of the genome due to the difficulty to sequence and assemble some regions (e.g., high GC content, large repeat regions, centromeres or telomeres). On the other hand, WES focuses on sequencing only the protein coding sequences. The primary advantages of WES are significantly reduced sequencing costs since less than 2% of the genome comprises exonic regions, the lower data storage costs, and more straightforward analyses (26). Whether it’s WES or WGS, data analysis remains the largest bottleneck in genomic studies, however the repertoire of available tools for sequence analysis is growing rapidly (27,28). Due to the error rates associated with the various NGS platforms, variants identified by NGS should be confirmed by Sanger sequencing technology (29,30).
Study design considerations
In addition to the importance of using the appropriate genomic tools for discovery of novel disease-causing variants, proper patient selection and phenotyping are an absolutely necessary component of study design. Team science approaches have proven critical in the study of numerous complex diseases and are increasingly emerging in the field of reproductive medicine.
Patient phenotyping
Careful phenotypic classification of infertile cases is of critical importance in the successful execution of genomic discovery studies. The identification of discrete and specific phenotypes has proven to yield early successes in the identification of novel genetic variants underlying male infertility. Two such examples are globozoospermia and MMAF. Globozoospermia is a specific defect of the sperm head, characterized by the absence of a sperm acrosome. Early genomic studies of both familial and sporadic cases successfully identified causal mutations in DPY19L2 and SPATA16 that cumulatively account for a large percentage of cases (31,32). Likewise, similar genomic analyses in men displaying MMAF have identified mutations in a number of genes including DNAH1, CFAP43, and CFAP44, among others (33,34). These cases illustrate the value of patient selection and phenotyping for genomic studies (8,35).
These examples are in contrast to the relatively slower-paced success in identifying genetic variants associated with spermatogenic impairment including oligozoospermia and NOA. While the formation of an acrosome or a competent flagellum during the late stages of spermiogenesis requires the concerted function of a relatively small number of genes, and disruption of either process yields a very specific phenotype, the process of spermatogenesis involves the concerted function of thousands of genes, and the disruption of any gene required for spermatogenesis can theoretically result in spermatogenic impairment.
This reality is reflected in the fact that, to date only a few high confidence variants have been discovered to explain severe spermatogenic impairment. Notably, more specific phenotypic characterization of NOA phenotypes based on testis histology data has proven to be important in variant discovery. Specifically, the genetic evaluation of patients with complete meiotic arrest, a relatively less common form of NOA, has produced a higher diagnostic rate than evaluation of the NOA population as a whole. More refined tools for molecular phenotyping of different categories of spermatogenic impairment or other classes of male infertility, such as single cell RNA sequencing will certainly improve patient classification and the diagnostic yield of genomic studies.
Family-based studies
The genomic analysis of families with two or more infertile siblings has proven to be an efficient and cost-effective tool for discovering high-confidence causal variants. In particular, families displaying increased consanguinity harbor a larger portion of the genome that is homozygous by descent, and thus are more likely to harbor homozygous recessive mutations. There are multiple examples in the literature of male infertility-causing mutations (29,30,36). Likewise, genomic analyses of non-consanguineous in which multiple siblings display the same infertility phenotype has proven successful. Screening both affected and unaffected siblings as well as both parents can reduce the number of potential variants to investigate many-fold. The continued identification of these families will be critical in identifying novel highly penetrant monogenic or associated variants.
Collaborative approaches
As medical research has evolved to integrate big data, population-scale approaches, team science has become absolutely imperative. This evolution has given rise to a large number of consortia focused on specific diseases and conditions. In recent years, the value of large-scale collaborations has become increasingly appreciated in the field of reproductive medicine as well. Two international genomics consortia [Genetics of Male Infertility Initiative (GEMINI; https://gemini.conradlab.org/)], and the International Male Infertility Genomics Consortium (IMIGC; http://www.imigc.org/)] have been organized in the past several years with the specific aim of organizing clinicians and basic researchers to enable the investigation of the genetic basis for various etiologies of male infertility. The fruits of these efforts are just beginning to emerge with several recent publications and numerous others in preparation or under review (30,37,38). In addition, the Male Reproductive Health Initiative (MRHI) has been organized to increase global awareness of male reproductive health and advocate to funding agencies the need for greater investment in research (39), and ReproUnion (https://reprounion.eu/) is organized to address big questions in reproductive medicine through international collaborations. Lastly, the Andrology Research Consortium (ARC) was founded to standardize data collection, clinical information and therapeutic approaches for the treatment of male infertility (40). As these and other efforts emerge and further mature, they will continue to accelerate the advancement of research in reproductive medicine.
Functional validation
While the identification of novel variants with plausible associations with male infertility is important, the functional validation of these variants is necessary to confirm variant function and to characterize the underlying mechanisms for infertility associated with specific variants. This is a huge undertaking considering the number of variants that are expected to underlie male infertility, however continuing improvements in genome editing tools, the emergence of novel research models and progress in the development of in vitro systems for the study of spermatogenesis will expand our ability to perform functional validation.
Animal models
The use of animal models for scientific purposes is both a longstanding practice in biological research, and a frequent matter of societal debate. However, this is an indisputably essential step to validate the results of a genetic study as well as to definitively establish a genetic marker, even though not all results obtained on animals can be directly translated to humans.
In reproduction, three different animal models are widely used to validate genomic discoveries: fruit fly, zebrafish and mice, due to their cost efficacy, easy maintenance, genetic homology and high fertility rates.
The fruit fly (Drosophila melanogaster) has been extensively studied for over a century as a model organism for genetic investigations because, on the molecular level, many similar features and pathways with humans were described. The main advantages are: short life cycle, ease of culture and maintenance, low number of chromosomes, small genome size (in terms of base pairs) and polytene chromosomes that permits a high level of gene expression. The main utility in reproductive studies is that Drosophila has a relatively short life span of 60–80 days, which makes it attractive for life span studies (41).
In the last two decades, zebrafish (Danio rerio) has emerged as a powerful model to study vertebrate development and disease because it has a short generation time, simplifying genetic manipulation and analysis, has a small size, has a high fecundity, and embryos are transparent. This last characteristic facilitates live imaging of developmental processes and make this animal model ideal to study infertility and reproduction (42).
Finally, mice (Mus musculus) are one of the most used animal models because of their phylogenetic relatedness and physiological similarity to humans and the ease of maintaining and breeding them in the laboratory. In addition, the availability of many inbred strains makes it ideal for conducting controlled genetic studies. Because mice are one of the most studied mammalian models and display many reproductive similarities with humans, mouse models have been described as “invaluable in dissecting the molecular mechanisms underlying a number of complex diseases like infertility” (43). Specifically, in rodent models it is worth recognizing that factors affecting male fertility may not adversely affect spermatogenesis, due to the highly efficient spermatogenesis seen in rodents, even though they may have a clinically important role in human male fertility (44).
Genetic manipulation
The generation of gene mutants in animal models, is necessary for demonstrating evidence for the role of a variant in male infertility. There are many tools available for the generation of gene mutants in animals. Here we describe a few including: N-ethyl-N-nitrosourea (ENU), morpholino oligonucleotides (MO), zinc finger nucleases (ZFN), Transcription activator-like effector nucleases (TALEN) and Clustered regularly interspaced short palindromic repeats (CRISPR/Cas) (Table 2).
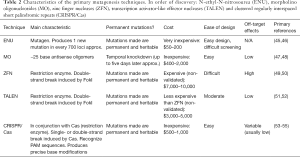
Full table
The ENU mutagenesis protocol is a very easy way to produce a mutant. ENU is an alkylating agent that transfers its ethyl group to nitrogen or oxygen radicals in DNA, resulting in base mispairing that causes base pair substitution. ENU is principally used when there is a need for highly efficient induction of point mutations randomly distributed throughout the germline (56). Therefore, the most interesting ENU application is the identification of novel alleles important in embryonic organogenesis. However, a genome-wide mutagenesis screen is mandatory to map the mutations generated by ENU (57).
On the other hand, MO is a type of antisense oligomer molecule designed to modify gene activity by blocking RNA translation or splicing (47). MOs are chemically synthesized, are similar to small interfering RNAs (siRNAs) and are typically injected into embryos at the 1-cell stage (58). MOs are typically 25-base oligomers with high specificity when designed appropriately. However, because the mechanism of action is based in gene expression modification instead of DNA modification, the duration of the effect is short-lived (typically 5–6 days), allowing the measure of the effects in early embryo development, but not later developmental stages. It is important to recognize that MO-induced phenotypes are often more severe than those of the corresponding mutants (59).
ZFN are the first generation of genome manipulation elements based on restriction enzymes (typically the restriction endonuclease FokI) (49). Zinc fingers are the most common DNA binding domain found in eukaryotes and are comprised of ~30 amino acids that interact with nucleotide triplets (each ZNF typically recognizes 3–6 nucleotide triplets). Zinc finger domains (usually 3 to 6) can be designed to target specific DNA sequences, and this enables zinc-finger nucleases to produce a double-strand brake (DSB) that promotes the nonhomologous DNA end joining (NHEJ) pathway activation. This technology can serve to induce random insertion-deletions (indels) when the NHEJ machinery works off-target or, using a supplied DNA fragment as a template for allele editing (50). Similarly, TALENs are based on restriction enzymes, however TALENs typically work with greater specificity and efficiency than ZFN. The main problem of ZFN and TALEN is that they can lead to toxicity or lethality due to binding at off-target sites resulting in the induction of undesired DNA cleavage (60). Even though TALEN design is generally more straightforward than ZNFs, to clone the large TALEN modules in series is challenging (51).
The latest gene editing technology is the CRISPR-Cas system discovered in the late 1980’s (53) but largely applied in gene editing only in the last 20 years (54,55). CRISPR systems are RNA-based bacterial defense mechanisms, primarily discovered in archaea organism, designed to recognize and eliminate foreign DNA from invading bacteriophage and plasmids. The system consists of a specific endonuclease, usually called Cas, that is directed to cleave a target sequence by a guide RNA (gRNA). Again, similar to the ZNF and TALEN systems, the CRISPR-Cas system can be used to either introduce specific mutations or insertions by co-injecting an engineered DNA construct or to introduce random mutations at the site of DNA cleavage by NHEJ machinery. In contrast to ZNF and TALEN, the design is easy, and the cost of this technology is relatively inexpensive (60).
In vitro systems
While animal models play a critical role in the validation of candidate mutations, there are limitations to the use of animal models. These include the cost of animal experiments, the time required for the development and completion of animal experiments, and the inherent genomic and reproductive differences between model organisms and humans. In many cases, genome sequences are not sufficiently conserved among species to accurately model specific mutations.
The culture of testicular tissues or cells in vitro to recapitulate spermatogenesis overcomes many of the shortcomings associated with model organism research, however in vitro culture of spermatogonial stem cells and in vitro spermatogenesis have not been achieved in humans in spite of success in mice (61,62) and tremendous efforts in humans (63). As a more complete understanding of human testis physiology and signaling cascades is gained, and as tools for three-dimensional culture of complex tissues continue to improve, in vitro spermatogenesis in humans will certainly be achieved. These advances coupled with CRISPR/Cas gene editing will enable the precise assessment of the impact of many variants on spermatogenesis. In addition, it will pave the way for the ability to restore spermatogenic capacity in vitro.
Beyond genetic markers for male infertility
Epigenetic modifications
Unlike the genome, the epigenome is highly variable between cells and is dynamic and can be influenced more readily by environmental and lifestyle factors. In the past two decades several studies have demonstrated that the sperm epigenome influences sperm function and fertilization. The basic epigenetic factors that exist in spermatozoa are histone and chromatin modifications, DNA methylation, and non-coding RNAs (ncRNAs) (64). Some authors suggest that the last two can be useful as epigenetic markers of male infertility.
DNA methylation
Numerous articles have described an association between male infertility and methylation abnormalities at particular imprinted loci in spermatozoa (65-68). Generally, these studies suggest that aberrations in the sperm DNA methylation epigenome can lead to defects throughout spermatogenesis, thereby impairing spermatogenesis or fertilization. However, the suggested link between sperm epigenetic changes and male sub-fertility is still a matter of debate, because the published studies are typically case-control analyses where the clinical relevance of the associations are uncertain.
In order to evaluate the clinical validity of some methylation variants, a recent meta-analytic analysis has been published and demonstrated that male infertility is associated with altered sperm DNA methylation at H19, MEST, and SNRPN imprinted genes, suggesting these as good candidate biomarkers of male infertility (69). H19 is one of the most studied imprinted gene for male infertility, encodes for a 2.3-kb non-coding mRNA and is strongly expressed during embryogenesis. This gene is located in chromosome 11 near the insulin-like growth factor 2 (IGF2) gene and is only expressed from the maternally inherited chromosome. This gene includes five exons and four introns and has different patterns of activation depending on the cell type and age (different between fetal life and adult life) suggesting that is involved in tissue differentiation (70). On the other hand, MEST encodes a member of the alpha/beta hydrolase superfamily and is located in chromosome 7. This gene is imprinted, and unlike H19, is monoallelically expressed from the paternal allele in fetal tissues (71). Finally, SNRPN is located within the Prader-Willi Syndrome critical region on chromosome 15 and is likewise expressed from the paternal allele. It encodes a component of the small nuclear ribonucleoprotein complex and may contribute to tissue-specific alternative splicing. This could be an important marker because some authors propose that ICSI predisposes to abnormal imprinting although there is not general consensus in this regard (72). More studies with these three epigenetic biomarkers are required to corroborate the associations suggested.
ncRNAs
Traditionally it was believed that the role of the male gamete was limited to the narrow window of fertilization as a simple vehicle for male DNA delivery to the embryo. However, several studies have demonstrated that this is not the case, and the male contribution, far from being confined to DNA, has been extended to a wide variety of molecules including coding and ncRNAs (73,74). Some studies have proposed that these transcripts are not just random remnants from early spermatogenesis stages but constitute a stable population that has been selectively retained, suggesting an important role in early zygotic development and postulating them as important infertility biomarkers (75).
Among ncRNAs, microRNAs (miRNAs) have been suggested as useful biomarkers of male infertility. Some authors suggested a panel of five well-known miRNAs in sperm (hsa-miR-34b*, hsa-miR-34b, hsa-miR-34c-5p, hsa-miR-429, and hsa-miR-122) as a novel noninvasive biomarker to diagnose patients with subfertility (76). However, in a recent study other authors suggested that pairs of miRNAs could be a more effective way to apply these epigenetic biomarkers, based on specificity and sensitivity values. Salas-Huetos and collaborators described the presence of 48 stable pairs of miRNAs in sperm of a fertile population of men (73) and suggested this group of miRNA pairs including miR-942-5p/miR-1208 and miR-34b-3p/miR-93-3p with a potential role in predicting fertility in a validation study (77).
Conclusions/future directions
Characterization of the underlying genetic basis for male infertility has long been a research focus for many groups. Early efforts focused on single gene screens by Sanger sequencing. These efforts successfully identified a handful of highly penetrant monogenic risk variants, and a large number of spurious associations. As genomic technologies have advanced, and whole genome approaches have been increasingly employed in the field, significant progress in understanding the genetic architecture of male infertility, as well as identifying novel highly penetrant monogenic risk variants, has been made.
To ensure continued progress in the field, future efforts should focus on the continued development of whole genome screens, improvements in analytical tools, novel phenotyping approaches, expanded sample sets, and advancements in in vitro and in vivo variant validation methods. In addition, the continued investigation of other factors such as epigenetics, lifestyle factors and exposures will improve our understanding of the etiologies of male infertility. Appropriately powered studies that will make significant strides in characterizing the genetic basis of male infertility can only be made through the combined efforts of scientists and clinicians collaborating across a broad spectrum of expertise. As these principles are increasingly employed, our understanding of the genetic basis for male infertility will likely expand rapidly in the coming years.
Acknowledgments
Funding: This work was supported, in part, by the National Institutes of Health (R01HD078641).
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editors (Keith Jarvi and Jared Bieniek) for the series “Genetic Causes and Management of Male Infertility” published in Translational Andrology and Urology. The article has undergone external peer review.
Conflicts of Interest: The authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tau.2020.03.43). The series “Genetic Causes and Management of Male Infertility” was commissioned by the editorial office without any funding or sponsorship. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Anderson JE, Farr SL, Jamieson DJ, et al. Infertility services reported by men in the United States: national survey data. Fertil Steril 2009;91:2466-70. [Crossref] [PubMed]
- World Health Organization. WHO laboratory manual for the examination and processing of human semen. Geneva: World Health Organization; 2010.
- Franken DR, Oehninger S. Semen analysis and sperm function testing. Asian J Androl 2012;14:6-13. [Crossref] [PubMed]
- Tomlinson MJ. Uncertainty of measurement and clinical value of semen analysis: has standardisation through professional guidelines helped or hindered progress? Andrology 2016;4:763-70. [Crossref] [PubMed]
- Ferlin A, Arredi B, Foresta C. Genetic causes of male infertility. Reprod Toxicol 2006;22:133-41. [Crossref] [PubMed]
- Krausz C, Riera-Escamilla A. Genetics of male infertility. Nat Rev Urol 2018;15:369-84. [Crossref] [PubMed]
- Tüttelmann F, Rajpert-De Meyts E, et al. Gene polymorphisms and male infertility--a meta-analysis and literature review. Reprod Biomed Online 2007;15:643-58. [Crossref] [PubMed]
- Oud MS, Volozonoka L, Smits RM, et al. A systematic review and standardized clinical validity assessment of male infertility genes. Hum Reprod 2019;34:932-41. [Crossref] [PubMed]
- Pan B, Li R, Chen Y, et al. Genetic association between androgen receptor gene CAG repeat length polymorphism and male infertility: A meta-analysis. Medicine (Baltimore) 2016;95:e2878 [Crossref] [PubMed]
- Luo H, Li H, Yao N, et al. Association between 3801T>C polymorphism of CYP1A1 and idiopathic male infertility risk: A systematic review and meta-analysis. PLoS One 2014;9:e86649 [Crossref] [PubMed]
- Zhang S, Tang Q, Wu W, et al. Association between DAZL polymorphisms and susceptibility to male infertility: Systematic review with meta-analysis and trial sequential analysis. Sci Rep 2014;4:4642. [Crossref] [PubMed]
- Ge YZ, Xu LW, Jia RP, et al. Association of polymorphisms in estrogen receptors (ESR1 and ESR2) with male infertility: A meta-analysis and systematic review. J Assist Reprod Genet 2014;31:601-11. [Crossref] [PubMed]
- Mobasseri N, Nikzad H, Karimian M. Protective effect of oestrogen receptor α-PvuII transition against idiopathic male infertility: a case-control study and meta-analysis. Reprod Biomed Online 2019;38:588-98. [Crossref] [PubMed]
- Wu Q, Zhang J, Zhu P, et al. The susceptibility of FSHB-211G>T and FSHR G-29A, 919A>G, 2039A>G polymorphisms to men infertility: An association study and meta-analysis. BMC Med Genet. BMC Med Genet 2017;18:81. [Crossref] [PubMed]
- Hong HH, Hu Y, Yu XQ, et al. Associations of C677T polymorphism in methylenetetrahydrofolate reductase (MTHFR) gene with male infertility risk: A meta-analysis. Eur J Obstet Gynecol Reprod Biol 2017;212:101-9. [Crossref] [PubMed]
- Dohle GR, Colpi GM, Hargreave TB, et al. EAU guidelines on male infertility. Eur Urol 2005;48:703-11. [Crossref] [PubMed]
- Punab M, Poolamets O, Paju P, et al. Causes of male infertility: A 9-year prospective monocentre study on 1737 patients with reduced total sperm counts. Hum Reprod 2017;32:18-31. [PubMed]
- Krausz C, Escamilla AR, Chianese C. Genetics of male infertility: from research to clinic. Reproduction 2015;150:R159-74. [Crossref] [PubMed]
- Huang T, Shu Y, Cai YD. Genetic differences among ethnic groups. BMC Genomics 2015;16:1093. [Crossref] [PubMed]
- Shinawi M, Cheung SW. The array CGH and its clinical applications. Drug Discov Today 2008;13:760-70. [Crossref] [PubMed]
- Yatsenko AN, Georgiadis AP, Röpke A, et al. X-linked TEX11 mutations, meiotic arrest, and azoospermia in infertile men. N Engl J Med 2015;372:2097-107. [Crossref] [PubMed]
- Hills A, Ahn JW, Donaghue C, et al. MLPA for confirmation of array CGH results and determination of inheritance. Mol Cytogenet 2010;3:19. [Crossref] [PubMed]
- Stuppia L, Antonucci I, Palka G, et al. Use of the MLPA Assay in the Molecular Diagnosis of Gene Copy Number Alterations in Human Genetic Diseases. Int J Mol Sci 2012;13:3245-76. [Crossref] [PubMed]
- Lima AC, Carvalho F, Gonçalves J, et al. Rare DMRT1 regulatory variants in severe spermatogenic failure. Andrology 2015;37:784-90.
- Robay A, Abbasi S, Akil A, et al. A systematic review on the genetics of male infertility in the era of next-generation sequencing. Arab J Urol 2018;16:53-64. [Crossref] [PubMed]
- Petersen BS, Fredrich B, Hoeppner MP, et al. Opportunities and challenges of whole-genome and -exome sequencing. BMC Genet 2017;18:14. [Crossref] [PubMed]
- Ke R, Mignardi M, Hauling T, et al. Fourth Generation of Next-Generation Sequencing Technologies: Promise and Consequences. Hum Mutat 2016;37:1363-7. [Crossref] [PubMed]
- Mignardi M, Nilsson M. Fourth-generation sequencing in the cell and the clinic. Genome Med 2014;6:31. [Crossref] [PubMed]
- Martinez G, Kherraf ZE, Zouari R, et al. Whole-exome sequencing identifies mutations in FSIP2 as a recurrent cause of multiple morphological abnormalities of the sperm flagella. Hum Reprod 2018;33:1973-84. [Crossref] [PubMed]
- Kasak L, Punab M, Nagirnaja L, et al. Bi-allelic Recessive Loss-of-Function Variants in FANCM Cause Non-obstructive Azoospermia. Am J Hum Genet 2018;103:200-12. [Crossref] [PubMed]
- Dam AH, Koscinski I, Kremer JA, et al. Homozygous mutation in SPATA16 is associated with male infertility in human globozoospermia. Am J Hum Genet 2007;81:813-20. [Crossref] [PubMed]
- Koscinski I, Elinati E, Fossard C, et al. DPY19L2 deletion as a major cause of globozoospermia. Am J Hum Genet 2011;88:344-50. [Crossref] [PubMed]
- Tang S, Wang X, Li W, et al. Biallelic Mutations in CFAP43 and CFAP44 Cause Male Infertility with Multiple Morphological Abnormalities of the Sperm Flagella. Am J Hum Genet 2017;100:854-64. [Crossref] [PubMed]
- Ben Khelifa M, Coutton C, Zouari R, et al. Mutations in DNAH1, which encodes an inner arm heavy chain dynein, lead to male infertility from multiple morphological abnormalities of the sperm flagella. Am J Hum Genet 2014;94:95-104. [Crossref] [PubMed]
- Bashamboo A, Mcelreavey KD. Male infertility. 1st ed. Rijeka, Croatia: InTech; 2012:204.
- Okutman O, Muller J, Baert Y, et al. Exome sequencing reveals a nonsense mutation in TEX15 causing spermatogenic failure in a Turkish family. Hum Mol Genet 2015;24:5581-8. [Crossref] [PubMed]
- Lopes AM, Aston KI, Thompson E, et al. Human Spermatogenic Failure Purges Deleterious Mutation Load from the Autosomes and Both Sex Chromosomes, including the Gene DMRT1. PLoS Genet 2013;9:e1003349 [Crossref] [PubMed]
- Lee AS, Rusch J, Lima AC, et al. Rare mutations in the complement regulatory gene CSMD1 are associated with male and female infertility. Nat Commun 2019;10:4626. [Crossref] [PubMed]
- De Jonge C, Barratt CLR. The present crisis in male reproductive health: an urgent need for a political, social, and research roadmap. Andrology 2019;7:762-8. [Crossref] [PubMed]
- Samplaski MK, Smith JF, Lo KC, et al. Reproductive endocrinologists are the gatekeepers for male infertility care in North America: results of a North American survey on the referral patterns and characteristics of men presenting to male infertility specialists for infertility investigations. Fertil Steril 2019;112:657-62. [Crossref] [PubMed]
- Markow TA. Drosophila reproduction: Molecules meet morphology. PNAS 2015;112:8168-9. [Crossref] [PubMed]
- Hoo JY, Kumari Y, Shaikh MF, et al. Zebrafish: A Versatile Animal Model for Fertility Research. Biomed Res Int 2016;2016:9732780 [Crossref] [PubMed]
- Tamowski S, Aston KI, Carrell DT. The use of transgenic mouse models in the study of male infertility. Syst Biol Reprod Med 2010;56:260-73. [Crossref] [PubMed]
- Johnson L, Ing NH, Welsh TH, et al. Efficiency of spermatogenesis in animals and humans. J Anim Sci 2000;77:1-47. [Crossref]
- Russell WL, Kelly E, Hunsicker P, et al. Specific-locus test shows ethylnitrosourea to be the most potent mutagen in the mouse. Proc Natl Acad Sci U S A 1979;76:5818-9. [Crossref] [PubMed]
- Lewis SE, Barnett LB, Shelby MD. ENU mutagenesis in the mouse electrophoretic specific-locus test 2. Mutational studies of mature oocytes. Mutat Res 1992;296:129-33. [Crossref] [PubMed]
- Summerton J. Morpholino antisense oligomers: the case for an RNase H-independent structural type. Biochim Biophys Acta 1999;1489:141-58. [Crossref] [PubMed]
- Partridge M, Vincent A, Matthews P, et al. A Simple Method for Delivering Morpholino Antisense Oligos into the Cytoplasm of Cells. Antisense Nucleic acid Drug Dev 1996;6:169-75. [Crossref] [PubMed]
- Miller J, Mclachlan AD, Klug A. Repetitive zinc-binding domains in the protein transcription factor LiA from Xenopus oocytes. EMBO J 1985;4:1609-14. [Crossref] [PubMed]
- Klug A. The Discovery of Zinc Fingers and Their Applications in Gene Regulation and Genome Manipulation. Annu Rev Biochem 2010;79:213-31. [Crossref] [PubMed]
- Boch J, Scholze H, Schornack S, et al. Breaking the Code of DNA Binding Specificity of TAL-Type III Effectors. Science 2009;326:1509-12. [Crossref] [PubMed]
- Bonas U, Stall RE, Staskawicz B. Genetic and Structural characterization of the avirulence gene avrBs3 from Xanthomonas campestris pv. vesicatoria. Mol Gen Genet 1989;218:127-36. [Crossref] [PubMed]
- Ishino Y, Shinagawa H, Makino K, et al. Nucleotide Sequence of the iap Gene, Responsible for Alkaline Phosphatase Isozyme Conversion in Escherichia coli, and Identification of the Gene Product. J Bacteriol 1987;169:5429-33. [Crossref] [PubMed]
- Mojica FJ, Díez-Villaseñor C, García-Martínez J, et al. Intervening Sequences of Regularly Spaced Prokaryotic Repeats Derive from Foreign Genetic Elements. J Mol Evol 2005;60:174-82. [Crossref] [PubMed]
- Mojica FJ, Juez G, Rodríguez-Valera F. Transcription at different salinities of Haloferax mediterranei sequences adjacent to partially modified Psfl sites. Mol Microbiol 1993;9:613-21. [Crossref] [PubMed]
- Olson E, Graham D. Animal Models in Pharmacogenomics. Handbook of Pharmacogenomics and Stratified Medicine, 2014:73-87.
- Stottmann R, Beier D. ENU Mutagenesis in the Mouse. Curr Protoc Hum Genet 2014;82:15.4.1-10.
- Summerton JE. Morpholino, siRNA, and S-DNA Compared: Impact of Structure and Mechanism of Action on Off-Target Effects and Sequence Specificity. Curr Top Med Chem 2007;7:651-60. [Crossref] [PubMed]
- Stainier DYR, Raz E, Lawson ND, et al. Guidelines for morpholino use in zebrafish. PLoS Genet 2017;13:e1007000 [Crossref] [PubMed]
- Gaj T, Gersbach CA, Iii CFB. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol 2013;31:397-405. [Crossref] [PubMed]
- Kita K, Watanabe T, Ohsaka K, et al. Production of Functional Spermatids from Mouse Germline Stem Cells in Ectopically Reconstituted Seminiferous Tubules1. Biol Reprod 2007;76:211-7. [Crossref] [PubMed]
- Sato T, Katagiri K, Gohbara A, et al. In vitro production of functional sperm in cultured neonatal mouse testes. Nature 2011;471:504-7. [Crossref] [PubMed]
- Ibtisham F, Wu J, Xiao M, et al. Progress and future prospect of in vitro spermatogenesis. Oncotarget 2017;8:66709-27. [Crossref] [PubMed]
- Bunkar N, Pathak N, Lohiya NK, et al. Epigenetics: A key paradigm in reproductive health. Clin Exp Reprod Med 2016;43:59-81. [Crossref] [PubMed]
- Kobayashi H, Sato A, Otsu E, et al. Aberrant DNA methylation of imprinted loci in sperm from oligospermic patients. Hum Mol Genet 2007;16:2542-51. [Crossref] [PubMed]
- Marques CJ, Carvalho F, Sousa M, et al. Genomic imprinting in disruptive spermatogenesis. Lancet 2004;363:1700-2. [Crossref] [PubMed]
- Marques CJ, Costa P, Vaz B, et al. Abnormal methylation of imprinted genes in human sperm is associated with oligozoospermia. Mol Hum Reprod 2008;14:67-74. [Crossref] [PubMed]
- Marques CJ, Francisco T, Sousa S, et al. Methylation defects of imprinted genes in human testicular spermatozoa. Fertil Steril 2010;94:585-94. [Crossref] [PubMed]
- Santi D, De Vincentis S, Magnani E, et al. Impairment of sperm DNA methylation in male infertility: a meta-analytic study. Andrology 2017;5:695-703. [Crossref] [PubMed]
- Gabory A, Ripoche MA, Yoshimizu T, et al. The H19 gene: regulation and function of a non-coding RNA. Cytogenet Genome Res 2006;113:188-93. [Crossref] [PubMed]
- Kosaki K, Kosaki R, Craigen WJ, et al. Isoform-Specific Imprinting of the Human PEG1/MEST Gene. Am J Hum Genet 2000;66:309-12. [Crossref] [PubMed]
- Manning M, Lissens W, Bonduelle M, et al. Study of DNA-methylation patterns at chromosome 15q11-q13 in children born after ICSI reveals no imprinting defects. Mol Hum Reprod 2000;6:1049-53. [Crossref] [PubMed]
- Salas-Huetos A, Blanco J, Vidal F, et al. New insights into the expression profile and function of micro-ribonucleic acid in human spermatozoa. Fertil Steril 2014;102:213-222.e4. [Crossref] [PubMed]
- Jodar M, Kalko S, Castillo J, et al. Differential RNAs in the sperm cells of asthenozoospermic patients. Hum Reprod 2012;27:1431-8. [Crossref] [PubMed]
- Krawetz SA. Paternal contribution: new insights and future challenges. Nat Rev Genet 2005;6:633-42. [Crossref] [PubMed]
- Abu-Halima M, Hammadeh M, Backes C, et al. A panel of five microRNAs as potential biomarkers for the diagnosis and assessment of male infertility. Fertil Steril 2014;102:989-997.e1. [Crossref] [PubMed]
- Corral-Vazquez C, Salas-Huetos A, Blanco J, et al. Sperm microRNA pairs: new perspectives in the search for male fertility biomarkers. Fertil Steril 2019;112:831-41. [Crossref] [PubMed]


