MiR-30b-3p and miR-126-3p of urinary extracellular vesicles could be new biomarkers for prostate cancer
Introduction
Prostate cancer (PCa) is the most common malignancy in elderly men in the United States, comprising 20% of all newly diagnosed male cancers in 2019 (1). An elevated level of prostate-specific antigen (PSA) and/or an abnormal digital rectal examination (DRE) can result in the performance of a prostate needle biopsy to diagnose potential PCa. However, the lack of specificity and sensitivity of PSA can result in unnecessary prostate biopsies; thus, new biomarkers for PCa need to be discovered. Urine is a non-invasively and easily collected source of potential new biomarkers, of which urinary markers of PCa such as PCA3 and the TMPRSS2-fusion gene have already been reported (2-6). In addition, urine collected following prostate massage can contain extracellular vesicles (EVs) secreted from PCa cells (7,8). EVs, including exosomes and microvesicles, are small vesicles secreted from various types of cells and are present in blood, urine, ascites, and saliva (9,10). EVs contain proteins, microRNAs (miRNAs), and messenger RNAs (mRNAs) and have an important role in intercellular communications between several different cell types. By communicating with other cells to induce angiogenesis and cancer progression, EVs originating from cancer cells promote the progression of cancer (11). Because proteins, miRNAs, and mRNAs in EVs may be indicators of the originating PCa cells, EVs could be potential sources from which new biomarkers for PCa can be discovered. Recently, proteins, mRNAs, and miRNAs in urinary EVs were reported as biomarkers of PCa (12-14). In the present study, EVs collected from post-DRE urine were analyzed by miRNA microarray, and we identified miRNAs that could be potential new biomarkers of PCa. We present the following article in accordance with the STARD 2015 reporting checklist (available at http://dx.doi.org/10.21037/tau-20-421).
Methods
Urine and serum samples
Urine and serum samples were collected from patients with moderately elevated serum PSA (4.0–25.0 ng/mL) examined at Osaka University Hospital between 2012 and 2017. All subjects gave their informed consent for inclusion before participating in this study, which was conducted in accordance with the Declaration of Helsinki. The study protocol was approved by the Osaka University Hospital Institutional Review Board (approval no. 13397-14). Serum and first-voided urine samples collected after DRE were initially stored at 4 °C for no more than 6 h before aliquoting and transferring them to storage at −80 °C until analysis. All patients also underwent transrectal ultrasound-guided biopsy.
EVs preparation and extraction of miRNA and mRNA of urinary EVs
Differential centrifugation was used to isolate EVs from 38.5 mL of the voided urine samples and 500 µL of serum samples. Cells were initially removed from the urine samples by centrifugation at 2,000 ×g for 30 min, following which the supernatant was centrifuged at 17,000 ×g for 30 min to remove debris and salts. After further ultracentrifugation of the supernatant at 130,000 ×g for 90 min and washing with PBS, the pellets underwent additional ultracentrifugation at 130,000 ×g for 90 min. The final obtained pellet was resuspended in 250 µL PBS and then stored at −80 °C for subsequent use. Patients’ sera were centrifuged at 300 ×g for 10 min to remove remaining cells and cell debris. After further ultracentrifugation of the supernatant at 150,000 ×g for 70 min and washing with PBS, the pellets underwent additional ultracentrifugation at 150,000 ×g for 70 min. The final obtained pellet was resuspended in 100 µL PBS and then stored at −80 °C for subsequent use. MiRNA and mRNA were isolated with a miRNeasy Mini Kit (Qiagen, Venlo, Netherlands), Clean-up Kit (Qiagen), and RNA MS2 (Roche Diagnostics, Mannheim, Germany) according to the manufacturers’ protocols.
Western blotting
The identity of urinary EVs was previously confirmed by the presence of the specific surface proteins CD9 and CD63 (15). Briefly, sodium dodecyl sulfate-polyacrylamide gel electrophoresis was used to separate 20 µL per well of original urine and urinary EVs. A semidry transfer system (Thermo Fisher Scientific, Waltham, MA, USA) was then used to transfer the gels to a polyvinylidene difluoride (PVDF: Thermo Fisher Scientific) membrane. The membranes were first probed with specific antibodies as indicated, followed by incubation with horseradish peroxidase-conjugated antibody against mouse immunoglobulin (1:1,000, Cell Signaling Technology, Beverly, MA, USA) and detection with enhanced chemiluminescence Western blotting detection reagents (Nacalai Tesque, Kyoto, Japan) using a ChemiDoc XRS Plus system (BIO-RAD) as the chemiluminescence detector. The antibodies used for immunological analysis in the present study were CD9 (1:1,000, 12A12, Shionogi, Osaka, Japan) and CD63 (1:1,000, ab59479, Abcam, Cambridge, UK).
Transmission electron microscopy
Ten-microgram samples of EVs were adsorbed onto a formvar/carbon-coated nickel grid for 1 h. The EVs were fixed with 2% paraformaldehyde and reacted with the anti-CD9 antibody. The immunoreactive EVs were then visualized with the anti-mouse IgG antibody preabsorbed with 20-nm gold particles. After negative staining with 2% aqueous uranyl acetate for 15 min, the samples were observed with a JEM-1400Plus transmission electron microscope (JEOL Ltd., Tokyo, Japan).
Microarray analysis of miRNAs from EVs
Microarray analysis for miRNAs of EVs was performed with EVs isolated from urine obtained after DRE from 14 men with elevated PSA [negative (n=4) and PCa of Gleason score (GS) 6 (n=3), GS 7 (n=3), and GS 8–9 (n=4)] on miRNA microarray 2.0 (Affymetrix, Santa Clara, CA, USA). An Affymetrix Gene Chip Scanner 3000 was used to scan the arrays, after which GeneSpring GX software (Agilent, Santa Clara, CA, USA) was used to process the scanned data.
Real-time polymerase chain reaction (RT-PCR)
Quantitative RT-PCR was performed to measure the expression of the miRNAs and mRNA, which were eluted from urinary EVs in 14 µL and 30 µL RNAase-free water, respectively. We used 3.75 µL of miRNA to synthesize 100 µL cDNA with a Mir-X miRNA First-Strand Synthesis Kit (Clontech, Mountain View, CA, USA) and 6 µL of mRNA to synthesize 400 µL cDNA with a Prime Script RT Reagent Kit (Clontech), according to the manufacturer’s instructions. MiRNA mapping was accomplished with miRBase 21. For RT-PCR of miRNA, a 20-µL reaction volume was prepared by mixing 1.6 µL cDNA, 10 µL SsoAdvanced Universal SYBR Green Supermix (BIO-RAD), 0.5 µL forward primer (10 µM), 0.5 µL reverse primer (10 µM), and 7.4 µL dH2O. Thermal cycling parameters were as follows: an initial step at 98 °C for 1 min followed by 40 cycles at 95 °C for 5 s and then 40 cycles at 66 °C for 15 s. The dissociation curves and melting temperatures were recorded. For RT-PCR of mRNA, a 20-µL reaction volume was prepared by mixing 2.0 µL cDNA, 10 µL SsoAdvanced Universal SYBR Green Supermix (BIO-RAD), 0.7 µL forward primer (10 µM), 0.7 µL reverse primer (10 µM), and 6.6 µL dH2O. Thermal cycling parameters were as follows: an initial step at 98 °C for 3 min, followed by 40 cycles at 95 °C for 15 s and 40 cycles at 60 °C for 30 s, with the dissociation curves and melting temperatures also recorded. Thermal Cycler CFX Connect (BIO-RAD) was used to perform the RT-PCR. To normalize miRNA expression in the urinary EVs, KLK3 in urinary EVs was used as the reference gene because KLK3 expression is specific to prostate tissue. The following oligonucleotides were used for RT-PCR analysis of KLK3 expression: KLK3 forward primer, 5'-CAGTCTGCGGCGGTGTT-3'; KLK3 reverse primer, 5'-GCAAGATCACGCTTTTGTTCCT-3'. PCR reactions were carried out in triplicate for each sample, and levels of miRNA expression in urinary EVs were calculated on the basis of the standard curve. The level of miRNA expression in serum EVs were calculated employing the comparative Ct method.
Statistical analysis
Patient characteristics were compared using the χ2 test and Mann-Whitney U test, which was also used for univariate analysis. Multiple regression analysis was used to assess the relative contributions of the factors of age and serum PSA and the candidate miRNAs in EVs. Logistic regression analysis was used to identify significant predictors of PCa. A P value <0.05 was considered to indicate statistically significant differences. Youden’s index was used to determine the optical cutoff value for each miRNA from the receiver-operator characteristics (ROC) curve, and the sensitivity and specificity to detect PCa with each miRNA were calculated according to each optical cutoff value. All statistical analyses were performed with JMP® Pro 13.0.0 (SAS Institute Inc, NC, USA).
The full study protocol can be accessed by contacting the corresponding author.
Results
EVs from human urine
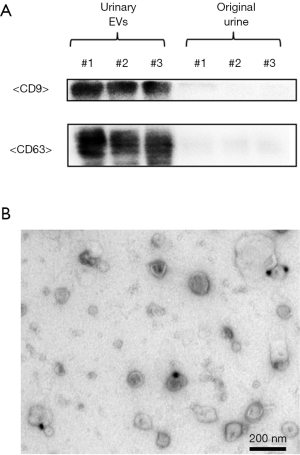
EVs were extracted by differential centrifugation from 38.5-mL samples of voided urine obtained following DRE and confirmed by Western blotting with the well-defined EV markers CD9 and CD63 (Figure 1A). Analysis by electron microscopy of EVs obtained from human urine following DRE showed rounded membrane-bound vesicles of <200 nm in size with CD9 expressed on their surface (Figure 1B).
Microarray analysis
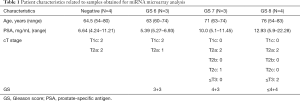
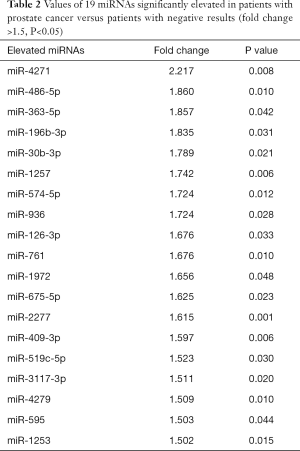
Patient characteristics related to the samples used in the microarray analysis are shown in Table 1. Microarray analysis showed that 19 miRNAs were significantly elevated in patients with PCa compared to patients with negative results (fold change >1.5, P<0.05, Table 2). After division of the patients into two groups [high-risk PCa group (GS 7–8) and low-risk PCa or negative group (GS 6 and negative)], no miRNAs were significantly elevated in the high-risk PCa group (fold change >1.5 and P<0.05).
Full table
Full table
Verification of biomarker candidates by RT-PCR
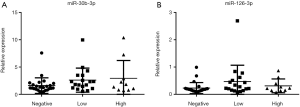
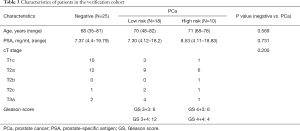
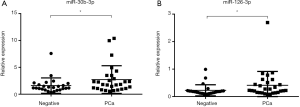
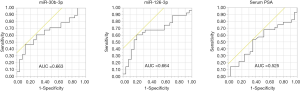
From the 19 identified miRNAs, we chose two (miR-30b-3p and miR-126-3p) as the candidate miRNAs because the other miRNAs could not be accurately measured in urinary EVs by RT-PCR due to multiple peaks in the dissociation curve. For verification as the independent cohort, 53 urine samples collected following DRE [25 samples from men with negative biopsy and 28 from men with PCa (6 men with GS 3+3, 12 with GS 3+4, 6 with GS 4+3, and 4 with GS 4+4)] were used (Table 3). Age, serum PSA, and clinical T stage were not significantly different between the negative and PCa patient groups (Table 3). miR-30b-3p and miR-126-3p were both significantly increased in the urinary EVs of men with PCa compared with those with a negative biopsy (P=0.0422 and 0.0405, respectively; Figure 2). We divided the PCa patients into two groups (those with GS 3+3/3+4 as the low-risk group and GS 4+3/4+4 as the high-risk group). Neither miR-30b-3p nor miR-126-3p was associated with either risk group (P for trend =0.102, 0.107, respectively; Figure 3). Analysis of the ROC curve showed the area under the curve (AUC) for the prediction of PCa by miR-30b-3p and miR-126-3p to be 0.663 [95% confidence interval (CI): 0.0114–0.805, P=0.0411] and 0.664 (95% CI: 0.0163–5.39, P=0.0477), respectively, whereas the AUC value was 0.525 (95% CI: −0.104–0.155, P=0.725) for prediction by serum PSA (Figure 4). The sensitivity and specificity of miR-30b-3p and miR-126-3p at the best cutoff values were 46.4% and 88.0% and 60.7% and 80.0%, respectively, and were better than those of serum PSA (53.5% and 64.0%, respectively). We also performed ROC curve analysis with the combination of miR-30b-3p and miR-126-3p. The AUC for the prediction of this model was 0.6657, but it was not significantly different from the model using only miR-30b-3p or miR-126-3p in urinary EVs (P=0.9827 and 0.9912, respectively).
Full table
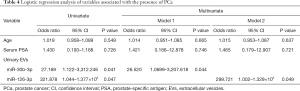
Finally, univariate logistic regression analysis showed both miR-30b-3p and miR-126-3p in EVs to be associated with the presence of PCa. After adjustment for age and serum PSA, the multivariate logistic regression analysis showed miR-30b-3p and miR-126-3p in urinary EVs to be significant predictors of the presence of PCa (Table 4). In addition, we investigated miR-30b-3p and miR-126-3p in serum EVs from 12 men with elevated PSA [negative (n=6] and PCa (n=6)]. There was no significant difference of miR-30b-3p and miR-126-3p in serum EVs between these two groups (Figure S1).
Full table
Discussion
PSA is hampered by its lack of specificity, and thus the discovery of new biomarkers for PCa are needed. In the present study, we studied urinary EVs in the discovery cohort, and identified 19 miRNAs in urinary EVs as candidate biomarkers of PCa. Furthermore, we analyzed urinary EVs in the independent cohort, and demonstrated that miR-30b-3p and miR-126-3p significantly elevated in patients with PCa compared to patients with negative results. Although neither miR-30b-3p nor miR-126-3p was associated with GS, the ROC analysis showed that the AUC for the prediction of PCa by miR-30b-3p and miR-126-3p were 0.663 and 0.664 respectively, which were better than that of PSA (AUC =0.525). These results showed that these miRNAs in urinary EVs could be new and better non-invasive biomarkers of PCa.
EVs are released from almost all types of cells into various bodily fluids, and cancer-specific EVs in bodily fluids could be a new biomarker. Urine is the promising source of biomarker discovery for the PCa. Because urine can be collected non-invasively and easily, and urine obtained after DRE includes abundant EVs derived from the prostate gland, urinary EVs after DRE could be an easily obtainable and specific biomarker for PCa. There was no difference of miR-30b-3p and miR-126-3p in serum EVs from the patients with or without PCa. Since EVs from many types of normal cells exist in the blood, the difference in the EV from the prostate might have been masked.
In the urinary EVs of the present study, we identified miR-30b-3p and miR-126-3p to be significant predictors of PCa in men with elevated PSA. Although neither miR-30b-3p nor miR-126-3p was associated with GS, the diagnostic performance of these miRNAs for PCa was better than that of serum PSA. MiR-30b-3p was reported to be a tumor suppressor in PCa (16). Kumar et al. reported that mir-30b-3p was downregulated in primary PCa and metastatic castration resistant PCa. They also showed that miR-30b-3p was positive regulator of androgen receptor signaling in PCa, and inhibition of miR-30b-3p enhanced the expression of androgen receptor and androgen-independent cell growth. MiR-126-3p was also reported to be a tumor suppressor in PCa (17). Hua et al. reported that miR-126 expression was significantly downregulated in PCa tissues and cell lines. They also showed that miR-126 inhibited PCa cell proliferation and metastasis, and reversed the epithelial-mesenchymal transition process in PCa (17).
There are only a few studies on miRNA in urinary EVs of PCa (14,18-23). Samsonov et al. first reported that miR-574, miR-141, and miR-21 expressions in urinary EVs were elevated in patients with PCa compared to healthy controls (14). ROC curve analysis of these miRNAs showed that the AUCs for the detection of PCa were 0.85, 0.86, and 0.65, respectively. However, the control group comprised healthy donors with a serum PSA below 4 ng/mL, and a comparison with serum PSA level was not carried out. Since then, some studies have reported the elevation of miRNAs such as miR-204, miR-21, miR-375, miR-145, and miR-2909, whereas others reported decreases in miRNAs such as miR-19b, miR-375, and miR-196a-5p in urinary EVs of PCa patients (18-23). However, in these reports the control group was healthy volunteer with normal serum PSA, or the cancer group included the patients with extremely elevated serum PSA (>100 ng/mL), or miRNAs were measured in only a single cohort and not verified in the independent cohort. To our knowledge this was the first report to identify miRNAs in urinary EVs among men with moderately elevated serum PSA (4.0–25.0 ng/mL) in the discovery cohort and verify in the independent cohort.
In our microarray analysis, we identified 19 miRNAs overexpressed in PCa patients compared to men with negative biopsy. However only two miRNAs were accurately measured in urinary EVs by RT-PCR. Some miRNAs could be measured only in cell-line lysate but not in human urinary EVs. One reason might be due to distinct isoforms (isomiRs), which were reported to be variations with respect to the reference miRNA sequence usually located at the 3’ end (24). A single miRNA hairpin arm has been shown to give rise to multiple isomiRs that are now referred to as mature miRNA transcripts and that can differ in length and sequence composition (25). The amounts of miRNAs including isomiRs were quantified in microarray experiments, but in RT-PCR experiments, the product derived from some isomiRs might result in multiple peaks in the dissociation curve and prevent accurate measurement.
There is also a study of the measurement of mRNA in urinary EVs by a commercially available test (ExoDx™). McKiernan et al. reported that a urinary 3-gene expression (PCA3, ERG, and SPDEF) assay predicted the presence of high-grade (GS ≥7) PCa in men ≥50 years of age with a PSA of 2–20 ng/mL (13). The AUC was 0.73 compared with that for PSA of 0.63. Recently, the result of ExoDx™ test in clinical setting was reported (26). Tutrone et al. conducted a prospective, randomised, blinded, two-armed clinical study among men aged ≥50 with PSA 2–10 ng/mL. Overall 68% of urologists decided to perform or differ prostate biopsies according to ExoDx™ test score. In result, 66% of high score patients had prostate biopsies, and only 26% of low score patients had prostate biopsies. ExoDx prostate use the urine without DRE, although it is known that DRE increases the biomarkers from the prostate in the urine (7,8). MiR-30b-3p and miR-126-3p might be detected in the urine without DRE in patients with PCa. Furthermore, the combination of microRNAs and PCA3, ERG and SPDEF might improve the detection of PCa. Further studied should be warranted.
There are several limitations in this study. First, it is a pilot study with a small sample size. Additional large-scale and multi-institutional studies should be performed to confirm our findings. Second, neither miR-30b-3p nor miR-126-3p was associated with GS. However, the diagnostic performance of these miRNAs for PCa was better than that of serum PSA. These miRNAs in urinary EVs could be useful to detect PCa in patients with an elevated PSA, thus reducing unnecessary biopsy. Third, the prognosis of PCa patients with high levels of miR-30b-3p and miR-126-3p in urinary EVs was not investigated. The prognostic performance of these miRNAs in urinary EVs should be studied in the future.
In conclusion, the present study suggested that miR-30b-3p and miR-126-3p in urinary EVs could be potential new biomarkers of PCa. Large-scale and multi-institutional studies will be required to further validate the present findings.
Acknowledgments
Funding: This work was supported by the Japan Society for the Promotion of Science (KAKENHI grant no. 19K09688).
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editor (Kazutoshi Fujita) for the series “Urinary Biomarkers of Urothelial Malignancies” published in Translational Andrology and Urology. The article was sent for external peer review organized by the editorial office.
Reporting Checklist: The authors have completed the STARD reporting checklist. Available at http://dx.doi.org/10.21037/tau-20-421
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tau-20-421
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tau-20-421). The series “Urinary Biomarkers of Urothelial Malignancies” was commissioned by the editorial office without any funding or sponsorship. KF served as the unpaid Guest Editor of the series and serves as an unpaid editorial board member of Translational Andrology and Urology from Oct 2019 to Sep 2021. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All subjects gave their informed consent for inclusion before participating in this study, which was conducted in accordance with the Declaration of Helsinki. The study protocol was approved by the Osaka University Hospital Institutional Review Board (approval no. 13397-14).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019;69:7-34. [Crossref] [PubMed]
- Fujita K, Ewing CM, Isaacs W, et al. Immunomodulatory IL-18 binding protein is produced by prostate cancer cells and its levels in urine and serum correlate with tumor status. Int J Cancer 2011;129:424-32. [Crossref] [PubMed]
- Okihara K, Ochiai A, Kamoi K, et al. Comprehensive assessment for novel prostate cancer markers in the prostate-specific antigen era: focusing on Asians and Asian countries. Int J Urol 2015;22:334-41. [Crossref] [PubMed]
- Fujita K, Hayashi T, Matsuzaki K, et al. Decreased fucosylated PSA as a urinary marker for high Gleason score prostate cancer. Oncotarget 2016;7:56643-9. [Crossref] [PubMed]
- Sanda MG, Feng Z, Howard DH, et al. Association between combined TMPRSS2:ERG and PCA3 RNA urinary testing and detection of aggressive prostate cancer. JAMA Oncol 2017;3:1085-93. [Crossref] [PubMed]
- Fujita K, Nonomura N. Urinary biomarkers of prostate cancer. Int J Urol 2018;25:770-9. [Crossref] [PubMed]
- Duijvesz D, Luider T, Bangma CH, et al. Exosomes as biomarker treasure chests for prostate cancer. Eur Urol 2011;59:823-31. [Crossref] [PubMed]
- Zijlstra C, Stoorvogel W. Prostasomes as a source of diagnostic biomarkers for prostate cancer. J Clin Invest 2016;126:1144-51. [Crossref] [PubMed]
- Urabe F, Kosaka N, Kimura T, et al. Extracellular vesicles: Toward a clinical application in urological cancer treatment. Int J Urol 2018;25:533-43. [Crossref] [PubMed]
- Hannafon BN, Ding WQ. Intercellular communication by exosome-derived microRNAs in cancer. Int J Mol Sci 2013;14:14240-69. [Crossref] [PubMed]
- Ciardiello C, Cavallini L, Spinelli C, et al. Focus on extracellular vesicles: new frontiers of cell-to-cell communication in cancer. Int J Mol Sci 2016;17:175. [Crossref] [PubMed]
- Fujita K, Kume H, Matsuzaki K, et al. Proteomic analysis of urinary extracellular vesicles from high Gleason score prostate cancer. Sci Rep 2017;7:42961. [Crossref] [PubMed]
- McKiernan J, Donovan MJ, O'Neill V, et al. Novel urine exosome gene expression assay to predict high-grade prostate cancer at initial biopsy. JAMA Oncol 2016;2:882-9. [Crossref] [PubMed]
- Samsonov R, Shtam T, Burdakov V, et al. Lectin-induced agglutination method of urinary exosomes isolation followed by mi-RNA analysis: Application for prostate cancer diagnostic. Prostate 2016;76:68-79. [Crossref] [PubMed]
- Matsuzaki K, Fujita K, Jingushi K, et al. MiR-21-5p in urinary extracellular vesicles is a novel biomarker of urothelial carcinoma. Oncotarget 2017;8:24668-78. [Crossref] [PubMed]
- Kumar B, Khaleghzadegan S, Mears B, et al. Identification of miR-30b-3p and miR-30d-5p as direct regulators of androgen receptor signaling in prostate cancer by complementary functional microRNA library screening. Oncotarget 2016;7:72593-607. [Crossref] [PubMed]
- Hua Y, Liang C, Miao C, et al. MicroRNA-126 inhibits proliferation and metastasis in prostate cancer via regulation of ADAM9. Oncol Lett 2018;15:9051-60. [Crossref] [PubMed]
- Koppers-Lalic D, Hackenberg M, de Menezes R, et al. Non invasive prostate cancer detection by measuring miRNA variants (isomiRs) in urine extracellular vesicles. Oncotarget 2016;7:22566-78. [Crossref] [PubMed]
- Foj L, Ferrer F, Serra M, et al. Exosomal and non-exosomal urinary miRNAs in prostate cancer detection and prognosis. Prostate 2017;77:573-83. [Crossref] [PubMed]
- Xu Y, Qin S, An T, et al. MiR-145 detection in urinary extracellular vesicles increase diagnostic efficiency of prostate cancer based on hydrostatic filtration dialysis method. Prostate 2017;77:1167-75. [Crossref] [PubMed]
- Wani S, Kaul D, Mavuduru RS, et al. Urinary-exosomal miR-2909: A novel pathognomonic trait of prostate cancer severity. J Biotechnol 2017;259:135-9. [Crossref] [PubMed]
- Bryzgunova OE, Zaripov MM, Skvortsova TE, et al. Comparative study of extracellular vesicles from the urine of healthy individuals and prostate cancer patients. PLoS One 2016;11:e0157566 [Crossref] [PubMed]
- Rodríguez M, Bajo-Santos C, Hessvik NP, et al. Identification of non-invasive miRNAs biomarkers for prostate cancer by deep sequencing analysis of urinary exosomes. Mol Cancer 2017;16:156. [Crossref] [PubMed]
- Morin RD, O'Connor M, Griffith M, et al. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res 2008;18:610-21. [Crossref] [PubMed]
- Lee LW, Zhang S, Etheridge A, et al. Complexity of the microRNA repertoire revealed by next-generation sequencing. RNA 2010;16:2170-80. [Crossref] [PubMed]
- Tutrone R, Donovan MJ, Torkler P, et al. Clinical utility of the exosome based ExoDx Prostate (IntelliScore) EPI test in men presenting for initial Biopsy with a PSA 2-10 ng/mL. Prostate Cancer Prostatic Dis 2020;23:607-14. [Crossref] [PubMed]