
Overexpression of MAX dimerization protein 3 (MXD3) predicts poor prognosis in clear cell renal cell carcinoma
Introduction
Kidney cancer is a common malignant tumor with more than 403,000 new cases diagnosed, and approximately 175,000 deaths are recorded every year worldwide (1). Renal cell carcinoma (RCC) originates from renal tubule epithelium and accounts for 90% of kidney cancers (2,3). Clear cell RCC (ccRCC) is the most frequent histological subtype of RCC, accounting for 80–90% of patients (4). The 5-year survival rate of localized ccRCC is about 65%, while metastatic ccRCC drops to 10–20% (5). ccRCC is recognized as an immunogenic and proangiogenic tumor, with immunotherapy and targeted therapy changing the treatment landscape for advanced RCC (6). Despite this, a substantial proportion of patients show no response to treatment due to innate or adaptive resistance (7). Although many driver gene mutations and epigenetic silencing of ccRCC have been discovered such as VHL, PBRM1, SETD2, and BAP1, drug resistance remains a significant barrier to treating ccRCC (8). For these reasons, exploring a molecular mechanism and identifying effective prognostic biomarkers in ccRCC is still imperative.
MAX dimerization protein 3 (MXD3) is a basic helix-loop-helix leucine zipper (bHLHZ) transcription factor affiliated with the MYC/MAX/MAD transcriptional network (9). The MAD family is composed of MXD1, MXI1, MXD3 and MXD4, which competes with MYC for heterodimerization with the cofactor MAX to bind DNA at E-box promoter sequences (9,10). MYC has been shown to promote cell cycle progression in many cell types, the MAD family known as “MYC antagonist” or “transcriptional repressor” inhibits cell proliferation (11). However, MXD3 behaves as an atypical member of the MAD family, as it functions more like MYC. MXD1, MXI1 and MXD4 are abundant in postmitotic cells, while MXD3 is detected in proliferating cells during the S-phase (12,13). MXD3 is overexpressed in many types of cancers and is required for medulloblastoma and acute lymphoblastic leukemia cell proliferation (14). The role of MXD3 in cancers is intriguing. In this study, we comprehensively analyzed the expression of MXD3 and its correlation with prognosis in ccRCC patients. These findings shed light on the important role of MXD3 in ccRCC and guide further preclinical research. We present the following article in accordance with the REMARK reporting checklist (available at: http://dx.doi.org/10.21037/tau-20-1187).
Methods
Data collection and differential expression analyses
The gene expression profiles and corresponding clinical information of ccRCC patients were downloaded from The Cancer Genome Atlas (TCGA) database (https://portal.gdc.cancer.gov/). This included 539 ccRCC samples and 72 adjacent normal kidney samples which were retrospectively studied. All samples were collected from patients diagnosed with ccRCC undergoing surgical resection. Patients were selected based on the criteria of TCGA, for which the requisites were that ccRCC was the primary tumor and that the patient had undergone no prior treatment, regardless of stage or histologic grade. Biospecimens were frozen and sufficiently sized. The adjacent normal kidney tissues were taken from greater than 2 cm away from the tumors. RNA was initially extracted from samples using a modification kit (Qiagen) (15).
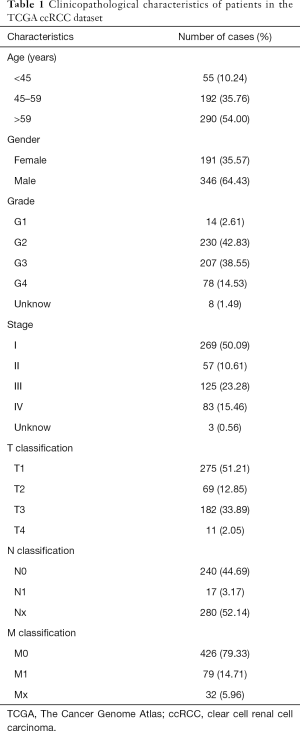
Clinical information of 537 patients was retrieved, including age, gender, histologic grade, pathological stage, tumor, node, metastasis (TNM) classification and follow-up information. The Mann-Whitney U test was used to compare the MXD3 expression levels between the 539 tumor samples and 72 adjacent normal kidney samples. The Wilcoxon matched-pairs test was applied to the analysis of paired samples. Statistical significance was set at P<0.05. Statistical analysis and graph plotting were performed using the SPSS Statistics 25 and GraphPad Prism 8 software packages. All procedures performed in this study were in accordance with the Declaration of Helsinki (as revised in 2013). and no ethical approval was required because the data we used were obtained from public databases. Because of the retrospective nature of the research, the requirement for informed consent was waived.
Survival analysis
To evaluate the relationship between the MXD3 expression and overall survival (OS), Kaplan-Meier survival analysis was performed using R version 3.6.1. OS was used as the primary endpoint and was recorded as the interval from diagnosis to death or the last follow-up date. The MXD3 expression median value was used as a cut-off to define the high expression and low expression groups. The log-rank test was used to test for differences in Kaplan-Meier survival probability.
Evaluation of associations between MXD3 expression and clinical parameters
We applied the Kruskal-Wallis test and Wilcoxon test to evaluate the associations between MXD3 expression and clinical characteristics using R version 3.6.1. Clinical factors related to OS were grouped. Patients were split into three groups based on age: “young” (less than 45 years), “middle-aged” (45 to 59 years), and “aged” (60 and up). Gender, histologic grade, pathological stage and TNM classification were grouped according to the corresponding standards.
Assessment of independent prognostic factors of OS for ccRCC
Univariate and multivariate Cox regression analysis were performed to assess the prognostic value of MXD3 expression, age, gender, histologic grade, pathological stage, and TNM classification in ccRCC. Before analysis, we examined the proportional hazards assumption. Data analysis and forest map plotting were conducted with SPSS Statistics 25, survival and survminer package in R. Statistical significance was set at P<0.05.
TIMER database analyses
To explore the correlation between MXD3 expression and tumor immune infiltration, TIMER database analysis was carried out. TIMER is a comprehensive web resource designed to estimate diverse immune cells’ clinical impact in different tumors (http://timer.cistrome.org/) (16). It applies a deconvolution method to infer the composition of tumor-infiltrating immune cells gene expression data (17). We used the TIMER database to investigate the correlation between MXD3 expression and the abundance of immune infiltrates, including B cells, macrophages, myeloid dendritic cells, CD4+ T cells, CD8+ T cells and neutrophils via immune association modules.
Methylation comparison and gene set enrichment analysis (GSEA)
To further explore the cause of the aberrant expression of MXD3 in ccRCC, we used the UALCAN database. UALCAN is a comprehensive resource for analyzing cancer data (http://ualcan.path.uab.edu/) and was used in this study to evaluate the epigenetic regulation of MXD3 expression by promoter methylation (18). To identify the underlying mechanism of MXD3 expression on ccRCC prognosis, we performed GSEA to determine statistically significant and concordant differences between the high and low expression groups of MXD3 using GSEA software version 4.1.0 (19). Gene set permutations were conducted 1,000 times with statistical significance set at P<0.05.
Results
MXD3 is overexpressed in ccRCC
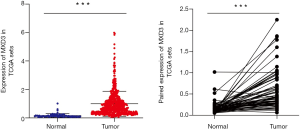
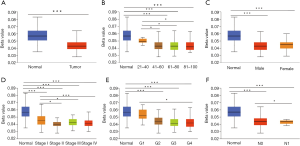
A total of 611 samples, including 539 ccRCC samples and 72 adjacent normal kidney samples from the TCGA database, were used in this study. The MXD3 expression values of tumor samples and adjacent normal samples did not comply with the normal distribution. The Mann-Whitney U test was used to compare the MXD3 expression levels of the 539 tumor samples with the 72 adjacent normal kidney samples. MXD3 was overexpressed in ccRCC significantly (P<0.001) (Figure 1A). There were 72 pairs of tumor tissues and matched adjacent normal tissues in the whole data. The MXD3 expression differences between 72 tumor samples and paired adjacent normal samples did not fit a normal distribution, and the Wilcoxon matched-pairs test was applied accordingly. MXD3 was also expressed higher in tumor tissues than matched adjacent normal tissues (P<0.001) (Figure 1B).
Overexpression of MXD3 is associated with poor prognosis in ccRCC
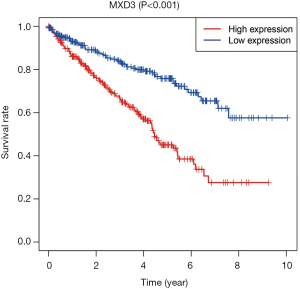
The 537 ccRCC patients with clinical information were divided into two groups according to MXD3 expression. The median expression value was used as a cut-off to define the high expression and low expression groups. The median follow-up time was 1,082 days. Kaplan-Meier survival analysis showed that patients with high MXD3 expression had significantly reduced OS compared to the low expression group (P<0.001) (Figure 2).
Associations between MXD3 expression and clinical features
Age, gender, clinical stage, histologic grade and TNM classification are important clinical features for cancer patients. Detailed clinical information for 537 ccRCC patients was available from the TCGA database (Table 1), and analysis of the associations between MXD3 expression and clinical features was conducted accordingly. MXD3 expression was significantly associated with histologic grade, clinical stage, T classification and M classification (P<0.001), while patients of different ages, genders, and N classifications showed no significant difference (Figure 3). MXD3 was expressed higher in patients of high grade, stage, T classification, and M classification. These findings suggested that ccRCC patients with high expression of MXD3 tended to progress to the advanced tumor stage more than patients with low MXD3 expression.
Full table

High expression of MXD3 is an independent risk factor for poor OS in ccRCC patients
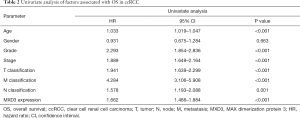
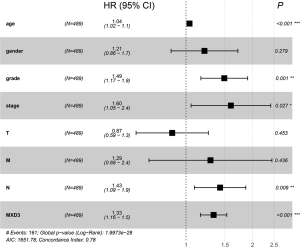
After the exclusion of 48 patients with incomplete clinical information, 489 patients were included in further Cox regression analysis. The univariate Cox analysis of factors affecting OS showed that high expression of MXD3, age, histologic grade, clinical stage, and TNM classification were associated with OS in ccRCC patients (Table 2). Multivariate Cox regression analysis confirmed that high expression of MXD3 was an independent risk factor for poor OS (P<0.001), along with age (P<0.001), histologic grade (P=0.001), clinical stage (P=0.027), and N classification (P=0.009) (Figure 4).
Full table
MXD3 expression is correlated with tumor-infiltrating immune cells
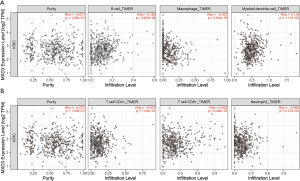
ccRCC is a type of immunogenic cancer and has an abundance of infiltrating immune cells. Infiltrating immune cells are generally considered beneficial for patients to destroy cancer cells and influence prognosis (6). Therefore, we studied the correlation between MXD3 expression and immune infiltration in ccRCC with the TIMER database. The results showed that MXD3 expression had no obvious correlation with tumor purity, however, MXD3 expression had significant positive correlations with B cells’ infiltrating levels and myeloid dendritic cells and negative correlation with macrophages (Figure 5A). There were no significant correlations between MXD3 expression and infiltrating levels of CD4+ T cells, CD8+ T cells and neutrophils (Figure 5B).
The MXD3 promoter region tends to be hypomethylated in ccRCC
To explore the reason for the high expression of MXD3 in ccRCC, the promoter methylation levels of MXD3 were analyzed. DNA methylation is an important epigenetic modification of the genome, regulating gene expression, and mediating epigenetic silencing in cancers and other diseases (20,21). Hypomethylation may induce chromosomal instability and gene activation (22). We studied the methylation levels of MXD3 promoter in ccRCC with the UALCAN database. The methylation levels of MXD3 promoter in ccRCC tended to be significantly hypomethylated compared with that in normal tissues (Figure 6A). ccRCC patients were also stratified according to age, gender, clinical stage, histologic grade and nodal metastasis. The results showed that patients of the aged, high clinical stage, high histologic grade and nodal metastasis had lower methylation levels than normal controls, however, promoter methylation levels did not change significantly in different gender subgroups (Figure 6B,C,D,E,F). This suggests that the promoter hypomethylation activate MXD3 expression in ccRCC.
GSEA identifies MXD3 related signaling pathways
To identify signaling pathways that were differentially activated between high and low MXD3 expression groups in ccRCC, we performed GSEA analysis. The results showed three significant Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways associated with high expression of MXD3, which were homologous recombination, base excision repair, and glycerophospholipid metabolism (Figure 7).

Discussion
ccRCC is the most common form of kidney cancer and has shown an increasing incidence in recent years (3). Approximately 20–40% of patients suffer late recurrence after curative surgery and patients with advanced ccRCC tended to have poor survival (23). Although immunotherapy and targeted therapy seem to yield a promising future for patients with ccRCC, a large proportion of patients receive no benefit from these therapies. The molecular mechanism of ccRCC has still not been elucidated completely and, until now, there has been no effective prognostic predictor for clinical practice. MXD3 is an atypical member of the MAD family, functioning more like MYC than an MYC antagonist. Within the canonical MYC/MAX/MXD network, the MAD family competes with MYC to heterodimerize with MAX to serve as transcription repressor. MXD3 is also found to be expressed in the S-phase of the cell cycle compared to other MAD family members in differentiated cells (24). The role of MXD3 in cell proliferation and tumorigenesis, along with its prognostic value in ccRCC is unclear. We performed comprehensive analyses integrating mRNA expression with clinical outcomes, tumor immune infiltration and promoter methylation to evaluate the value of MXD3 in ccRCC.
In this study, we analyzed the expression levels of MXD3 and prognostic influence in ccRCC. The aberrant expression levels of MXD3 between tumor and normal tissues have been observed in several cancers. MXD3 expression has been observed to be elevated in glioblastoma, medulloblastoma, neuroblastoma, hepatocellular carcinoma and precursor B-cell acute lymphoblastic leukemia (14,25-28). MXD3 is recommended as a prognostic and progression biomarker for hepatocellular carcinoma (27). We compared MXD3 expression in ccRCC with normal tissues using data from the TCGA database. The data sets showed that MXD3 expression was markedly higher in tumor tissues than normal tissues on the mRNA level. The paired comparison method was also applied, and the results were consistent. The Kaplan-Meier curve for OS showed that overexpression of MXD3 predicts poor prognosis. Univariate Cox analysis indicated that high expression of MXD3 was a potential independent prognostic factor for ccRCC. Multivariate Cox analysis confirmed the prognostic value of MXD3. Age, histologic tumor grade, clinical stage, and N classification were also presented to be independent risk factors for OS of ccRCC patients by multiple Cox analyses, which was in accordance with previous studies (29). Molecular markers can improve the accuracy of prognostic models (30). MXD3 shows valuable clinical application prospect in the prognostic prediction of ccRCC.
There was no significant difference in MXD3 expression among different groups divided by age, gender, or nodal metastasis, however, we found that MXD3 expression was associated with histologic grade, clinical stage, T classification and M classification. The MXD3 expression increased as the tumor progressed to higher levels. It has been reported that MXD3 promotes granule neuron precursor proliferation and is also necessary for medulloblastoma cell proliferation (25,31). Human precursor B acute lymphoblastic leukemia cells are also shown to be enhanced in apoptosis with loss of MXD3 (32). The selective expression pattern and potential role in tumorigenesis of MXD3 indicate that it may serve as a novel therapy target. MXD3 siRNA nanocomplexes have been developed to treat neuroblastoma and precursor B-cell acute lymphoblastic leukemia (26,28,33). MXD3 antisense oligonucleotides show promising prospect to be a new therapeutic regimen in specific cancers of the central nervous system and hematologic system. Drugs of this type could induce side effects such as cytotoxicity or inflammatory response because a fraction of nanoparticles or their remnants remains in cells. The cytotoxicity may be reduced with the degradation of the remnants or by modification of the nanoparticles. Modified antisense oligonucleotides nanocomplexes can be delivered intracellularly without major toxicity, and when combined with conventional chemotherapy it can also reduce dosage and lessen risks of chemotherapy drugs. Considering the characteristic gene expression and prognostic influence of MXD3 in ccRCC, MXD3 may be a novel therapeutic target for ccRCC.
RCC stands out as an abundant immune-infiltrated tumor, and the tumor microenvironment profoundly affects the response to therapy (34). We investigated the correlations between MXD3 expression and diverse immune infiltration levels in ccRCC. The results revealed positive relationships between MXD3 expression and infiltration of B cells and myeloid dendritic cells, significant negative correlations between infiltration of macrophages and MXD3 expression. Although much research regarding tumor immunity is focused on T cells, research around B cells’ role in tumors is still in the nascent period. B cells can positively regulate antitumor immunity by producing antibodies, cytokines, and acting as antigen-presenting cells, while the immunosuppressive effects of B regulatory cells are increasingly appreciated (35). The absence of B cells could enhance the antitumor response (36). Dendritic cells are versatile and potent antigen-presenting cells to induce T cells and B cells responses (37). Macrophages show a dual role in tumor progression that express suppress or promote functional programs by M1/M2 polarization (38). The correlations between MXD3 expression and immune cell infiltration suggest that MXD3 may influence the recruitment and regulation of infiltrating immune cells and indicate the potential mechanism whereby MXD3 regulates tumor microenvironment in ccRCC.
MXD3 has been demonstrated to be a transcriptional target of E2F1 (39). However, the causes of high expression of MXD3 in ccRCC have not been clarified. We investigated the methylation level in ccRCC and found that the methylation levels of MXD3 promoter in tumor tissues were significantly reduced compared with normal controls. Gene expression may be activated due to hypomethylation of gene promoter, the elevated MXD3 expression may be explained by its hypomethylation. The methylation levels of aged patients were lower than those seen in young patients, which was in consistent with the results of previous studies (40). There was no significant difference in methylation between female and male groups, which was in accordance with MXD3 expression levels. Patients with higher clinical stage, histologic grade, and N classification showed lower methylation levels, which confirmed the rationality of hypomethylation up-regulating MXD3 expression in ccRCC.
GSEA identified three important signaling pathways between high and low MXD3 expression groups: homologous recombination, base excision repair, and glycerophospholipid metabolism. Homologous recombination and base excision are important biological behaviors for accessing genetic information in the context of cell proliferation (41,42). Glycerophospholipid metabolism is an important metabolic activity in cells, which not only provides basic components for cellular structure but also functions in signaling transduction and cancers (43). Considering MXD3 is preferentially expressed in the S-phase of the cell cycle, all results indicate that MXD3 may play important roles in the cell proliferation of ccRCC.
Conclusions
This study comprehensively analyzed the differential expression of MXD3 between ccRCC and normal kidney tissues. Overexpression of MXD3 was correlated with poor prognosis and high expression of MXD3 was associated with hypomethylation of MXD3 promoter in ccRCC. MXD3 may play important roles in cell proliferation and immune infiltration of ccRCC, and the underlying mechanism will be verified in further research. In conclusion, MXD3 shows promising clinical prospect and could be an effective prognostic biomarker and potential therapeutic target for ccRCC.
Acknowledgments
We are grateful to The Cancer Genome Atlas (TCGA) and the Clinical Biobank of Beijing Hospital for providing public data in this study.
Funding: None.
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at http://dx.doi.org/10.21037/tau-20-1187
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tau-20-1187
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tau-20-1187). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in this study were in accordance with the Declaration of Helsinki (as revised in 2013) and no ethical approval was required because the data we used were obtained from public databases. Because of the retrospective nature of the research, the requirement for informed consent was waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. Erratum in: CA Cancer J Clin. 2020 Jul;70(4):313. doi: 10.3322/caac.21609. Epub 2020 Apr 6. [Crossref] [PubMed]
- Hsieh JJ, Purdue MP, Signoretti S, et al. Renal cell carcinoma. Nat Rev Dis Primers 2017;3:17009. [Crossref] [PubMed]
- Znaor A, Lortet-Tieulent J, Laversanne M, et al. International variations and trends in renal cell carcinoma incidence and mortality. Eur Urol 2015;67:519-30. [Crossref] [PubMed]
- Ljungberg B, Albiges L, Abu-Ghanem Y, et al. European Association of Urology Guidelines on Renal Cell Carcinoma: The 2019 Update. Eur Urol 2019;75:799-810. [Crossref] [PubMed]
- Fendler A, Bauer D, Busch J, et al. Inhibiting WNT and NOTCH in renal cancer stem cells and the implications for human patients. Nat Commun 2020;11:929. [Crossref] [PubMed]
- Liu XD, Hoang A, Zhou L, et al. Resistance to Antiangiogenic Therapy Is Associated with an Immunosuppressive Tumor Microenvironment in Metastatic Renal Cell Carcinoma. Cancer Immunol Res 2015;3:1017-29. [Crossref] [PubMed]
- Martin CJ, Datta A, Littlefield C, et al. Selective inhibition of TGFβ1 activation overcomes primary resistance to checkpoint blockade therapy by altering tumor immune landscape. Sci Transl Med 2020;12:eaay8456. [Crossref] [PubMed]
- Hakimi AA, Voss MH, Kuo F, et al. Transcriptomic Profiling of the Tumor Microenvironment Reveals Distinct Subgroups of Clear Cell Renal Cell Cancer: Data from a Randomized Phase III Trial. Cancer Discov 2019;9:510-25. [Crossref] [PubMed]
- Grandori C, Cowley SM, James LP, et al. The Myc/Max/Mad network and the transcriptional control of cell behavior. Annu Rev Cell Dev Biol 2000;16:653-99. [Crossref] [PubMed]
- Ayer DE, Kretzner L, Eisenman RN. Mad: a heterodimeric partner for Max that antagonizes Myc transcriptional activity. Cell 1993;72:211-22. [Crossref] [PubMed]
- Hurlin PJ, Huang J. The MAX-interacting transcription factor network. Semin Cancer Biol 2006;16:265-74. [Crossref] [PubMed]
- Quéva C, Hurlin PJ, Foley KP, et al. Sequential expression of the MAD family of transcriptional repressors during differentiation and development. Oncogene 1998;16:967-77. [Crossref] [PubMed]
- Fox EJ, Wright SC. S-phase-specific expression of the Mad3 gene in proliferating and differentiating cells. Biochem J 2001;359:361-7. [Crossref] [PubMed]
- Ngo T, Corrales A, Bourne T, et al. Alternative Splicing of MXD3 and Its Regulation of MXD3 Levels in Glioblastoma. Front Mol Biosci 2019;6:5. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature 2013;499:43-9. [Crossref] [PubMed]
- Li T, Fu J, Zeng Z, et al. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res 2020;48:W509-14. [Crossref] [PubMed]
- Li T, Fan J, Wang B, et al. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res 2017;77:e108-10. [Crossref] [PubMed]
- Chandrashekar DS, Bashel B, Balasubramanya SAH, et al. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017;19:649-58. [Crossref] [PubMed]
- Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545-50. [Crossref] [PubMed]
- Heyn H, Esteller M. DNA methylation profiling in the clinic: applications and challenges. Nat Rev Genet 2012;13:679-92. [Crossref] [PubMed]
- Keshet I, Schlesinger Y, Farkash S, et al. Evidence for an instructive mechanism of de novo methylation in cancer cells. Nat Genet 2006;38:149-53. [Crossref] [PubMed]
- Feinberg AP, Tycko B. The history of cancer epigenetics. Nat Rev Cancer 2004;4:143-53. [Crossref] [PubMed]
- Brookman-May SD, May M, Shariat SF, et al. Time to recurrence is a significant predictor of cancer-specific survival after recurrence in patients with recurrent renal cell carcinoma--results from a comprehensive multi-centre database (CORONA/SATURN-Project). BJU Int 2013;112:909-16. [Crossref] [PubMed]
- Hooker CW, Hurlin PJ. Of Myc and Mnt. J Cell Sci 2006;119:208-16. [Crossref] [PubMed]
- Barisone GA, Ngo T, Tran M, et al. Role of MXD3 in proliferation of DAOY human medulloblastoma cells. PLoS One 2012;7:e38508. [Crossref] [PubMed]
- Duong C, Yoshida S, Chen C, et al. Novel targeted therapy for neuroblastoma: silencing the MXD3 gene using siRNA. Pediatr Res 2017;82:527-35. [Crossref] [PubMed]
- Xu B, Lv W, Li X, et al. Prognostic genes of hepatocellular carcinoma based on gene coexpression network analysis. J Cell Biochem 2019. Epub ahead of print. [Crossref] [PubMed]
- Satake N, Duong C, Chen C, et al. Targeted therapy with MXD3 siRNA, anti-CD22 antibody and nanoparticles for precursor B-cell acute lymphoblastic leukaemia. Br J Haematol 2014;167:487-99. [Crossref] [PubMed]
- Gu L, Li H, Wang Z, et al. A systematic review and meta-analysis of clinicopathologic factors linked to oncologic outcomes for renal cell carcinoma with tumor thrombus treated by radical nephrectomy with thrombectomy. Cancer Treat Rev 2018;69:112-20. [Crossref] [PubMed]
- Klatte T, Rossi SH, Stewart GD. Prognostic factors and prognostic models for renal cell carcinoma: a literature review. World J Urol 2018;36:1943-52. [Crossref] [PubMed]
- Yun JS, Rust JM, Ishimaru T, et al. A novel role of the Mad family member Mad3 in cerebellar granule neuron precursor proliferation. Mol Cell Biol 2007;27:8178-89. [Crossref] [PubMed]
- Barisone GA, Satake N, Lewis C, et al. Loss of MXD3 induces apoptosis of Reh human precursor B acute lymphoblastic leukemia cells. Blood Cells Mol Dis 2015;54:329-35. [Crossref] [PubMed]
- Yoshida S, Duong C, Oestergaard M, et al. MXD3 antisense oligonucleotide with superparamagnetic iron oxide nanoparticles: A new targeted approach for neuroblastoma. Nanomedicine 2020;24:102127. [Crossref] [PubMed]
- Vuong L, Kotecha RR, Voss MH, et al. Tumor Microenvironment Dynamics in Clear-Cell Renal Cell Carcinoma. Cancer Discov 2019;9:1349-57. [Crossref] [PubMed]
- Zhang Y, Gallastegui N, Rosenblatt JD. Regulatory B cells in anti-tumor immunity. Int Immunol 2015;27:521-30. [Crossref] [PubMed]
- Tadmor T, Zhang Y, Cho HM, et al. The absence of B lymphocytes reduces the number and function of T-regulatory cells and enhances the anti-tumor response in a murine tumor model. Cancer Immunol Immunother 2011;60:609-19. [Crossref] [PubMed]
- Worbs T, Hammerschmidt SI, Förster R. Dendritic cell migration in health and disease. Nat Rev Immunol 2017;17:30-48. [Crossref] [PubMed]
- Tamura R, Tanaka T, Yamamoto Y, et al. Dual role of macrophage in tumor immunity. Immunotherapy 2018;10:899-909. [Crossref] [PubMed]
- Fox EJ, Wright SC. The transcriptional repressor gene Mad3 is a novel target for regulation by E2F1. Biochem J 2003;370:307-13. [Crossref] [PubMed]
- Horvath S, Raj K. DNA methylation-based biomarkers and the epigenetic clock theory of ageing. Nat Rev Genet 2018;19:371-84. [Crossref] [PubMed]
- Wright WD, Shah SS, Heyer WD. Homologous recombination and the repair of DNA double-strand breaks. J Biol Chem 2018;293:10524-35. [Crossref] [PubMed]
- Madders EC, Parsons JL. Base Excision Repair in Chromatin and the Requirement for Chromatin Remodelling. Adv Exp Med Biol 2020;1241:59-75. [Crossref] [PubMed]
- Gonzalez-Baro MR, Coleman RA. Mitochondrial acyltransferases and glycerophospholipid metabolism. Biochim Biophys Acta Mol Cell Biol Lipids 2017;1862:49-55. [Crossref] [PubMed]


