
Development and validation of prognostic nomograms and a web-based survival rate calculator for sarcomatoid renal cell carcinoma in pre- and post-treatment patients
Introduction
Renal cell carcinoma (RCC) accounts for 2–3% of all adult malignancies, for which the incidence increases annually (1). Clear cell, papillary, and chromophobe RCC account for the majority of diagnoses, while other histological subtypes with type-specific clinical and pathological features and prognosis also present. Variation in the impact of pathological subtypes is not considered in the current RCC prognostic models included in the American Joint Committee on Cancer (AJCC) and University of California Los Angeles Integrated Staging Systems (UISS) (2).
Sarcomatoid renal cell carcinoma (SRCC) is a rare variant of RCC that comprises approximately 5% of cases. Moreover, SRCC arises from any subtype of epithelial RCC (e.g., clear cell, papillary, or chromophobe) and presents as a deeply dedifferentiated tumor (3). SRCC is also associated with a poor clinical outcome and advanced clinicopathological features (4). Currently, there are few studies on SRCC, and the difference in prognosis between traditional renal carcinoma and SRCC remains unclear. In addition, there is no specific accepted prognostic model for SRCC. Cases were retrieved from the National Cancer Institute’s Surveillance, Epidemiology, and End Results (SEER) database to construct prognostic nomograms and a web-based survival rate calculator for SRCC. We present the following article in accordance with the TRIPOD reporting checklist (available at http://dx.doi.org/10.21037/tau-20-1192).
Methods
Data source and study population
A cohort of SRCC patients diagnosed from 2004 to 2015 was drawn from the SEER database, which incorporates high-quality data derived from 18 cancer registries and covers approximately 27.8% of the U.S. population based on the 2010 census (5). The SRCC ICD-O-3 8318 code includes both sarcomatoid and conventional RCC with Kidney Parenchyma CS Site-Specific Factor 4 code 010. ICD-O-3 8310 includes conventional RCC and combines the histologic types of clear cell adenocarcinoma, NOS [8312], papillary adenocarcinoma, NOS [8260], RCC [8317], chromophobe type, and adenocarcinoma with mixed subtypes [8255]. Only microscopically confirmed RCC patients were included. The pathology of patients who did not undergo surgery was identified and diagnosed by a needle biopsy. Cases without follow-up or vital status and those with Kidney Parenchyma CS tumor size code 000, and the absence of a mass/tumor were also excluded. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Since identifiable patient information is not contained in the publicly available SEER database, no ethical approval was required.
Patient variables
Patient data was extracted from SEER database fields, which included “age”, “race”, “gender”, “histologic type ICD-O-3”, “derived AJCC stage group, 6th ed. (2004+)”, “derived AJCC T, 6th ed. (2004+)”, “derived AJCC N, 6th ed. (2004+)”, “derived AJCC M, 6th ed. (2004+)”, “RX Summ-Surg Prim Site (1998+)”, “CS tumor size (2004+)”, “CS site-specific factor 4 (2004+)”, “SEER cause-specific death classification”, “survival months”, and “vital status recode (study cutoff used)”.
Nomogram construction
The SEER SRCC dataset was divided into a training cohort (70%) and a validation cohort (30%) for model evaluation using R version 3.5.3 (available from: http://www.r-project.org/) and the caret package (available from: http://topepo.github.io/caret/index.html). A multivariate Cox regression analysis of the overall survival (OS) and cancer-specific survival (CSS) of the training cohorts identified the independent prognostic factors used to construct nomograms predicting the patients’ 3- and 5-year OS and CSS. The inclusion criteria in the nomogram included patient age, sex, ethnicity, TNM stage, tumor size, and whether surgery was performed. The analysis was performed with the rms, foreign, and survival packages of R version 3.5.3. A final model was selected via a backward stepdown process using the Akaike information criterion (6).
Validation of nomograms
Nomogram discrimination (i.e., specificity and sensitivity) was estimated and calibration was performed with R version 3.5.3 statistics and the rms, foreign, survival, and survival receiver-operating characteristic (ROC) packages. The OS and CSS derived from the nomograms and from the AJCC staging system were compared using R and the rcorrp.cens (Hmisc) package to calculate the C-index. Nomogram discrimination was determined by the area under the ROC curve (AUC). Calibration curves were plotted so that the predictive rates fell on a 45º diagonal line. A random resampling bootstrapping procedure using 1,000 resamples was used for internal validation. The nomograms were validated using the SEER validation cohort.
Decision curve analysis
While traditional diagnostic test indicators (e.g., sensitivity, specificity, and area under the ROC curve) only measure the diagnostic accuracy of the predictive model and fail to consider the clinical utility of a particular model, the advantage of a DCA is that it integrates the preferences of patients or decision-makers into the analysis. This concept meets the practical needs of clinical decision-making and has been increasingly applied in the analysis of clinical samples. Here, we compared the nomogram with two other models commonly used in clinical practice, one of which is the more frequently used sixth edition of AJCC staging, whereas the other is T staging to clarify the superiority of the nomogram.
Web construction
The age at diagnosis was stratified as <62, 62–76, and >76 by X-tile, while the tumor size was stratified into three parts as <5.5, 5.5–13.5, and >13.5. X-tile can select the optimal cut-off value in the statistical process. The “shiny”, “devtools”, and “Dyn-Nom” packages were used to generate a web-based survival rate calculator via shiny project in R studio. The calculator predicted the survival of patients according to their clinical characteristics. The default survival time was the median OS month, which could be selected by sliding according to the predicted needs. The specific operation method was to select the patient conditions in the selection box on the left, and then select the prediction time, so that the OS or CSS could be observed from the figure on the right. Moreover, the Numerical Summary was used to view the results of the detailed forecast data.
Statistical analysis
The values of unordered categorical variables were compared using a chi-square test. Ordered categorical variables were compared by Goodman and Kruskal’s gamma. If the expected frequency was less than five, a Fisher’s exact test was used. A Bonferroni’s correction was used for multiple comparisons. Continuous variables were compared with a Student’s t-test. The Mann-Whitney U test was used to compare variables that did not have a normal distribution. Cumulative survival was estimated using the Kaplan-Meier method and compared with log-rank tests. The statistical analysis was performed with SPSS 25.0 for Windows (IBM Corp., Armonk, NY, USA). All tests were two sided and a P value <0.05 was considered significant.
Results
Data
A total of 3,670 SRCC cases consisted of 1,894 cases with ICD-O-3 code 8318 and 1,776 cases of conventional RCC cases with Kidney Parenchyma CS Site-Specific Factor 4 code 010 were identified in the SEER database; 10 cases with a CS tumor size code of 0 were deleted before selection, 923 cases without information in the T/N/M stage, and 5 cases without survival data were excluded. The remaining 2,742 SRCC cases were included in the analysis and divided into a training cohort of 1,921 cases for model development and validation cohort of 821 cases for model evaluation. The specific inclusion and screening criteria are presented in Figure 1. The median follow-up times were 13 months for both the SEER training and validation cohorts. Table 1 presents the clinical characteristics of the two cohorts.
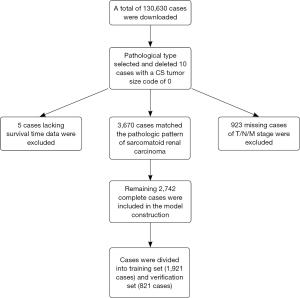
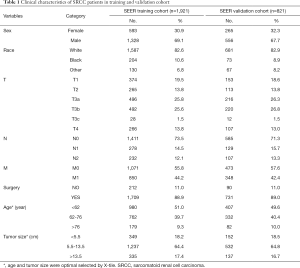
Full table
Variables independently associated with the OS and CSS are shown in Table 2. A multivariate Cox regression analysis of the training cohort found that age, race, sex, T stage, N stage, M stage, surgery or no surgery, and tumor size were independently associated with the OS; age, T stage, N stage, M stage, surgery or no surgery, and tumor size were independently associated with CSS.
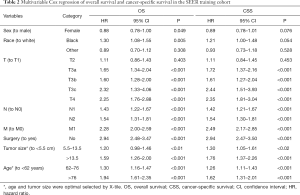
Full table
Prognostic nomograms and validation
In the SEER training cohort, the 3-year OS was 22.2%, 5-year OS was 11.0%, and the median OS was 14 months. The 3-year CSS was 44.8%, 5-year CSS was 24.3%, and median CSS was 32 months. Among all of the cases, there were 1,816 overall deaths, of which 1,654 deaths were specifically related to disease. The prognostic nomograms integrated all the independent prognostic factors for the OS and CSS in the training cohort and are shown in Figure 2. The final nomograms were simplified from preliminary versions, which integrated the 7th or 8th edition AJCC TNM N1 and N2 staging criteria. Surgery could not be confirmed in three of the SRCC training cohort cases; however, since their characteristics were nearly identical to cases with confirmed surgery, they were included with the analysis of surgical cases.
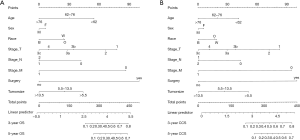
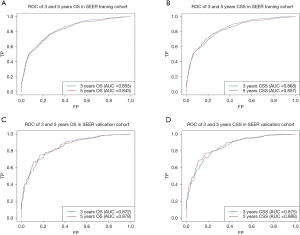
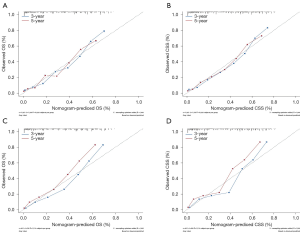
The AUC of the nomograms was 0.855 for predicting the 3-year OS and 0.843 for the 5-year OS (Figure 3A) and 0.868 for the 3-year CSS and 0.857 for the 5-year CSS (Figure 3B). The AUC of the SEER nomogram validation cohort was 0.872 for predicting the 3-year OS and 0.878 for the 5-year OS (Figure 3C). It was 0.875 for the 3-year CSS and 0.888 for the 5-year CSS (Figure 3D). The nomograms displayed moderate discrimination. The calibration curves for the probability of the 3- or 5-year OS and CSS predicted by the nomograms were in agreement with the actual observations (Figure 4A,B), while the calibration curves for the probability of the 3- and 5-year OS and CSS of the SEER validation cohort predicted by the nomograms were in agreement with clinical observations (Figure 4C,D). The accuracy of the nomograms for predicting the OS and CSS in the training cohort were more accurate than that of the AJCC stage system. The C-indexes for the nomogram predictions were 0.767 (95% CI: 0.753–0.780) for the OS and 0.775 (95% CI: 0.761–0.788) for the CSS, which were both significantly higher (P<0.01) than the AJCC sixth edition staging of 0.725 (95% CI: 0.705–0.745 for the OS and 0.715 (95% CI: 0.695–0.735) for the CSS.
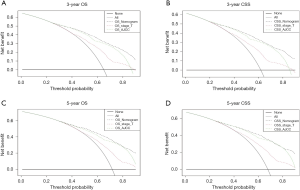
The results of the DCA were demonstrated by the 3-year OS and CSS (Figure 5A,B) as well as the 5-year OS and CSS (Figure 5C,D). The decision analysis curve provided a better net benefit than the other two evaluating systems at the same probability threshold. In the comparison of the different models in the decision analysis curves, we observed the advantages of the nomograms. The results showed that both the 3- or 5-year OS and CSS were obviously superior to the other two models in comparison.
Web-based survival rate calculator
According to the constructed nomogram, we established a dynamic web-based calculator (available from: https://yangtong.shinyapps.io/specific/) to predict the CSS and (available from: https://yangtong.shinyapps.io/overall/) to predict the OS of patients with primary SRCC according to a nomogram. With the web-based survival rate calculator that we built, we were better able to evaluate our patients in the clinic, and thus contribute to better treatment. For example, the 5-year OS probability was approximately 39.0% (95% CI: 31.1–48.0%) and the 5-year specific survival was approximately 41.0% (95% CI: 34.0–51.0%) for white male patients aged younger than 62 years with stage T3aN1M0, accepted surgery, and a tumor size less than 5.5 cm.
Discussion
SRCC consists of a mixture of malignant elements, including RCC and transitional cell carcinoma. RCC has been regarded as exhibited sarcomatoid transformation when it harbors a population of atypical spindle cells that resemble spindle cell sarcoma. Sarcomatoid transformation has been recognized as a pattern of “dedifferentiation” with a loss of the characteristic epithelial features of RCC (7). However, the criteria for defining SRCC vary from series to series, with the majority of studies requiring the identification of an epithelial component within the tumor. The proportion of spindle cells necessary to permit the designation of a tumor as SRCC varies considerably. This variability in the definition contributes to considerable inter-study differences in the proportion of a different series of renal tumors. The presence of sarcomatoid features in the RCC is known to be an independent predictor of poor survival (8-10); however, few available prognostic models are specific for SRCC. Zhang et al. published a multivariate prognostic model based on the outcomes of 204 post-nephrectomy SRCC patients (11). In addition, Gu et al. developed a nomogram to predict the OS of SRCC based on 103 post-nephrectomy patients (12). Both models were derived using data from small patient cohorts treated at a single center and both lack external validation. Data from the SEER database was used to develop and validate prognostic nomograms for SRCC. The SEER database is a large population-based dataset that may reflect real-world conditions with a higher accuracy. The developed and verified nomogram model has filled the gap in the lack of survival prognostic models in the field of sarcoma -type renal cancer research. Compared with the survival models of other disease types, this rare pathological type is more difficult to come by due to a fewer number of patients. For patients with sarcoma type renal cancer that is poorly differentiated and associated with a poor prognosis, diagnosis is often associated with a later stage. Therefore, it is crucial to evaluate whether patients will benefit from surgery and how to better guide clinical treatment.
The SEER data may be limited by unrecorded variables, underreported and incomplete data, variations in data coding and reporting, and the migration of patients in and out of the SEER registry. The main limitation of the SRCC data is a lack of information regarding the percentage of cases with sarcomatoid characteristics. Therefore, the influence of a different proportion of sarcoma tissues on patient prognosis was unclear. Previous studies have demonstrated that a greater percentage of sarcomatoid is associated with a worse outcome (13). Other information (e.g., margin status) cannot be downloaded from the database, which may affect the patient prognosis. In addition, it is unfortunate that the specific metastatic organs and the number of metastases are unknown among patients with stage M1. If we can understand and analyze patients with distant metastasis in detail, we will refine our model and facilitate new thinking about the prognosis of patients with advanced tumors. The results may be affected by the short follow-up period of the patients and the overall small sample size, which may affect the results. Furthermore, different therapeutic drugs and surgical procedures were not included in the SEER databases, which may affect the survival time.
Nomograms have been shown to be more accurate than the conventional staging systems for predicting the prognosis in many other cancers (14,15). The nomograms constructed in this study to predict the 3- and 5-year OS and CSS for SRCC performed well in both the SEER training and validation cohorts. The nomograms displayed better predictive accuracy for survival compared with the 6th edition of the AJCC staging system. After the AJCC stage system has been updated and the TNM system in seventh and eighth editions published, changes in the N and T stages, especially the T3 stage, should be taken into consideration when applying the developed nomograms (16). In the constructed nomogram model, we noted that patients with an older age, larger tumors, and without surgery had lower scores, and there was a shorter corresponding survival time. However, in the T stage, we noticed that patients the in T3c stage had a poorer prognosis than that of the T4 patients, which may be related to the shedding of inferior vena cava embolus and accompaniment of the occurrence of cardiac and brain accidents. This may also be caused by a sample deviation caused by the small number of patients in T3c.
By constructing a network calculator, we were able to observe the survival outcomes of patients in different states of operability. Moreover, we could preoperatively determine the impact of undergoing surgery on the survival of a certain patient. In clinical practice, whether a patient receives surgical treatment is dependent on the patient’s comprehensive situation (e.g., whether there is a combination of other diseases and surgical contraindications, and nutritional status). Therefore, the present model provides an additional reference for clinical decision-making.
Conclusions
Prognostic nomograms with sufficient specificity for predicting the OS and CSS of pre- and post-treatment SRCC patients were developed and validated with data from the SEER database. Therefore, a web-based survival rate calculator can be used to intuitively predict the survival possibility of SRCC patients.
Acknowledgments
We thank International Science Editing (http://www.internationalscienceediting.com) for editing this manuscript.
Funding: This study was supported by financial grants from the National Natural Science Foundation of China (grant No. 81902355), Shandong Provincial Natural Science Foundation (grant No. ZR2018MH024), and the Focused Research and Development Program of Shandong Province (grant No. 2017GSF18130).
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at http://dx.doi.org/10.21037/tau-20-1192
Peer Review File: Available at http://dx.doi.org/10.21037/tau-20-1192
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tau-20-1192). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Ethical approval was waived by the local ethics committee, as SEER data is publicly available and de-identified. Informed consent was not required because personal identifying information was not involved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Patard JJ, Kim HL, Lam JS, et al. Use of the University of California Los Angeles integrated staging system to predict survival in renal cell carcinoma: an international multicenter study. J Clin Oncol 2004;22:3316-22. [Crossref] [PubMed]
- Shuch B, Bratslavsky G, Linehan WM, et al. Sarcomatoid renal cell carcinoma: a comprehensive review of the biology and current treatment strategies. Oncologist 2012;17:46-54. [Crossref] [PubMed]
- Zhang L, Wu B, Zha Z, et al. The prognostic value and clinicopathological features of sarcomatoid differentiation in patients with renal cell carcinoma: a systematic review and meta-analysis. Cancer Manag Res 2018;10:1687-703. [Crossref] [PubMed]
- Surveillance, Epidemiology, and End Results (SEER) Program () SEER*Stat Database: Incidence - SEER 18 Regs Research Data + Hurricane Katrina Impacted Louisiana Cases, Nov 2018 Sub (1975-2016 varying) - Linked To County Attributes - Total U.S., 1969-2017 Counties, National Cancer Institute, DCCPS, Surveillance Research Program, released April 2019, based on the November 2018 submission.www.seer.cancer.gov
- Harrell FE Jr, Lee KL, Mark DB. Multivariable prognostic models: issues in developing models, evaluating assumptions and adequacy, and measuring and reducing errors. Stat Med 1996;15:361-87. [Crossref] [PubMed]
- Mouallem NE, Smith SC, Paul AK. Sarcomatoid renal cell carcinoma: Biology and treatment advances. Urol Oncol 2018;36:265-71. [Crossref] [PubMed]
- Cheville JC, Lohse CM, Zincke H, et al. Sarcomatoid renal cell carcinoma: an examination of underlying histologic subtype and an analysis of associations with patient outcome. Am J Surg Pathol 2004;28:435-41. [Crossref] [PubMed]
- Shuch B, Said J, La Rochelle JC, et al. Cytoreductive nephrectomy for kidney cancer with sarcomatoid histology--is up-front resection indicated and, if not, is it avoidable? J Urol 2009;182:2164-71. [Crossref] [PubMed]
- Holz S, Tosco L, Akand M, et al. Pushing the limits of metastasis-directed treatment in metastatic renal cell carcinoma in the era of targeted therapy. Urol Oncol 2020;38:937.e1-937.e9. [Crossref] [PubMed]
- Zhang BY, Thompson RH, Lohse CM, et al. A novel prognostic model for patients with sarcomatoid renal cell carcinoma. BJU Int 2015;115:405-11. [Crossref] [PubMed]
- Gu L, Ma X, Li H, et al. Prognostic value of preoperative inflammatory response biomarkers in patients with sarcomatoid renal cell carcinoma and the establishment of a nomogram. Sci Rep 2016;6:23846. [Crossref] [PubMed]
- de Peralta-Venturina M, Moch H, Amin M, et al. Sarcomatoid differentiation in renal cell carcinoma: a study of 101 cases. Am J Surg Pathol 2001;25:275-84. [Crossref] [PubMed]
- Ehdaie B, Shariat SF, Savage C, et al. Postoperative nomogram for disease recurrence and cancer-specific death for upper tract urothelial carcinoma: comparison to American Joint Committee on Cancer staging classification. Urol J 2014;11:1435-41. [PubMed]
- Balachandran VP, Gonen M, Smith JJ, et al. Nomograms in oncology: more than meets the eye. Lancet Oncol 2015;16:e173-80. [Crossref] [PubMed]
- Merrill MM, Wood CG, Tannir NM, et al. Clinically nonmetastatic renal cell carcinoma with sarcomatoid dedifferentiation: Natural history and outcomes after surgical resection with curative intent. Urol Oncol 2015;33:166.e21-9. [Crossref] [PubMed]


