
Eef2k is not required for fertility in male mice
Introduction
Infertility is estimated to impact 8–12% of couples worldwide, and is defined by a failure to conceive following unprotected frequent sexual intercourse over a one-year period. Approximately half of infertile couples are impacted by male-factor infertility (1), which can occur due to hormonal deficits (2), physical causes, sexually transmitted diseases, or environmental, lifestyle (3,4), and genetic factors (5,6). As spermatogenesis is a key process in the maintenance of appropriate male fertility, any abnormalities in the generation of spermatozoa can give rise to male-factor infertility (7). There are four primary phases of spermatogenesis: (I) the initial mitotic proliferation and spermatogonial differentiation of pre-leptotene spermatocytes; (II) the meiotic division of spermatocytes to yield spermatids; (III) spermiogenesis wherein round spermatids are converted into elongating spermatids; and (IV) spermiation, wherein elongated spermatids, as called as sperm, are released into the lumen of seminiferous tubules (8).
We have previously shown that Tcte1-knockout male mice are sterile as they exhibit sperm with impaired progressive motility (asthenozoospermia) (9). In our quantitative proteomic analyses, we found eukaryotic elongation factor-2 kinase (Eef2k) to be significantly downregulated in these Tcte1-knockout mice, suggesting that Eef2k may be an important regulator of spermatogenesis and/or sperm motility.
Eef2k is a structurally and functionally atypical kinase involved in calmodulin-mediated signaling that functions by phosphorylating and inhibiting Eef2 to regulate protein translation and synthesis (10,11). Eef2k overexpression has previously been reported in breast, pancreatic, brain, and lung cancers in which it plays important roles in the regulation of apoptosis, autophagy, and cell cycle progression, making it a viable target for anticancer treatment (12-16). However, the in vivo functionality of Eef2k in the testes remains to be clarified. As such, we herein used a CRISPR/Cas9 approach to generate Eef2k-knockout mice, and we used these mice to explore the role of Eef2k in testicular development and spermatogenesis. We present the following article in accordance with the ARRIVE reporting checklist (available at http://dx.doi.org/10.21037/tau-21-18).
Methods
Eef2k-knockout mouse generation
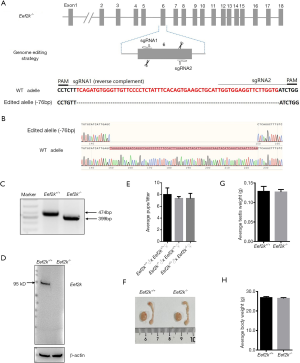
The study was conducted in accordance with the declaration of Helsinki (as revised in 2013). Experiments were performed under a project license (No.: IACUC-2004020) granted by the Animal Ethical and Welfare Committee, in compliance with the Institutional Animal Care and Use Committee of Nanjing Medical University guidelines. To generated targeted knockout mice, Cas9 mRNA and single-guide RNAs (sgRNAs) were generated as in prior studies (7,9,17,18). Briefly, AgeI and DraI were used for the respective linearization of the Cas9 and sgRNA plasmids, which were subsequently purified using the MinElute PCR Purification Kit (Qiagen, Duesseldorf, Germany). An mMESSAGE mMACHINE T7 Ultra Kit (Ambion, TX, USA) was used to facilitate the in vitro transcription of Cas9 mRNA, followed by purification with the RNeasy Mini Kit (Qiagen) based on provided instructions. Similarly, sgRNAs were generated and purified with the MEGA shortscript and MEGA clear kits (Ambion), respectively. The sgRNAs used in the present study were prepared to target Eef2k exon 6, and were as follows: 5'-GGGGAACAACCCACATCTGAAAGAGG-3' and 5'-TGGTGGAGGTTCTTGGTGATCTGG-3'. These two constructs were then injected into murine zygotes that were obtained following the mating of WT male and super-ovulated female C57BL/6 mice. The strain is stable and easy to reproduce. The pure background C57BL/6 mice for gene knockout can ensure the high stability of genetic background and the consistency of experimental data.
Genotyping
Edited Eef2k founders were identified via PCR amplification (Rapid Taq master mix, Vazyme, Nanjing, China) using the following primers: F 5'-GGAGATGGTGATCTTCATACT-3' and R 5'-GCATACTATTCTGTGGTAGGA-3', and were subcloned into the pMD19-T plasmid (TaKaRa, Wuhan, China), after which standard Sanger sequencing was performed. Those founders that exhibited Eef2k frameshift mutations were mated with WT C57BL/6 mice for a minimum of three generations to ensure that no off-target editing was present in these animals, yielding respective WT and knockout Eef2k alleles that were 474 bp and 399 bp in length. Sanger resequencing of Eef2k-/- mice was conducted, with results being plotted using SnapGene (version 1.1.3). Tail biopsy-derived DNA samples were subjected to PCR amplification with appropriate primers and were separated via agarose gel electrophoresis for genotyping analyses.
Fertility analyses
Fertility testing in male mice was conducted by mating each male mouse with three WT C57BL/6 females. Females were evaluated for the presence of a vaginal plug each morning, after which birth dates and numbers of pups in each litter were assessed.
Western blotting
RIPA buffer (P0013C, Beyotime, Shanghai, China) supplemented with protease inhibitors (B14002, Bimake, TX, USA) was used to prepare protein lysates, which were subsequently separated via 10% SDS-PAGE and transferred to PVDF membranes. Blots were blocked using 5% non-fat milk in TBST for 2 h. Blots were incubated overnight at 4 °C with anti-EEF2K (ab45168; 1:1,000; Abcam, China) and anti-β-actin (ac026; 1:10,000; ABclonal, Wuhan, China). Blots were then washed thrice with TBST (5 min/wash), after which they were probed for 2 h with secondary antibodies (1:1,000) at room temperature (19). The High-sig ECL Western Blotting Substrate (Tanon, Shanghai, China) was used to detect protein bands.
Histological analyses
Davidson’s fluid was used to fix murine epididymis or testes tissues overnight, after which the tissues were transferred into 70% ethanol. Samples were subsequently dehydrated with an ethanol gradient (80%, 90%, 100%) and were paraffin-embedded. Tissue sections (5 µm-thick) were then prepared, deparaffinized using dimethylbenzene, and testes and epididymis sections were respectively subjected to periodic acid Schiff (PAS) and hematoxylin and eosin (H&E) staining (20,21). Additionally, slides were smeared with sperm samples from the cauda epididymis, which were subsequently fixed for 30 min using 4% paraformaldehyde in PBS, after which samples were rinsed and subjected to H&E staining.
Immunofluorescence and TUNEL staining
Tissue sections and sperm samples were deparaffinized, rehydrated, and antigen retrieval was conducted in sodium citrate buffer by boiling samples for 10 min. Samples were cooled and blocked with 5% BSA in PBS for 2 h, followed by overnight probing with primary antibodies (Table S1) at 4 °C. Samples were then washed thrice using PBST (0.05% Tween 20 in 1× PBS), probed with secondary antibodies (Table S2), and counterstained at room temperature with Hoechst 33342 (1:1,000; Invitrogen, CA, USA) for 2 h. After an additional wash, sections were mounted, and apoptotic cells were detected via the terminal deoxynucleotidyl transferase dUTP nick end-labeling (TUNEL) assay (Vazyme) based upon provided directions (22,23). An LSM800 confocal microscope (Carl Zeiss AG, Jena, Germany) was used to image and evaluate samples.
Computer-assisted sperm analysis
Sperm samples were collected by generating cross-incisions throughout the cauda epididymis and extruding the cells therein and resuspending them in human tubal fluid culture medium (In Vitro Care, MD, USA). Individual 10 µl sperm samples were subjected to a computer-assisted semen analysis (CASA; Hamilton-Thorne Research, Inc, MA, USA) of motility, progressive motility, and sperm concentrations (24).
Eepididymal sperm count
Mature sperm from homozygous and WT mice were obtained by making small incisions throughout the cauda epididymis, followed by extrusion and suspension in culture medium, after dilution, count with blood cell counting plate under microscope. The sperm on the grid line are included in the corresponding grid according to the position of the head of sperm.
Statistical analysis
Data were analyzed using Microsoft Excel and GraphPad Prism 6.0 via one-way ANOVAs or unpaired two-tailed t-tests. All experiments were repeated three or more times, and data were given as means ± SD. P<0.05 was the significance threshold for this study.
Results
Eef2k−/− mouse generation
To explore the functional importance of Eef2k in the testes of C57BL/6 mice, Eef2k-/-were prepared via a CRISPR/Cas9 approach (Figure 1A). A frameshift mutation was introduced into Eef2k by deleting a 76 bp portion of exon 6 of this gene, as confirmed via Sanger sequencing and PCR genotyping (Figure 1B,C). We were not able to detect any Eef2k protein in the testes of our male Eef2k−/− mice (Figure 1D), and these mice exhibited normal viability and body weight relative to their WT counterparts.
Eef2k−/− mice exhibited normal fertility and spermatogenesis
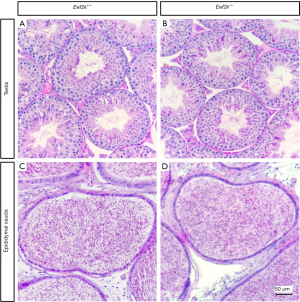
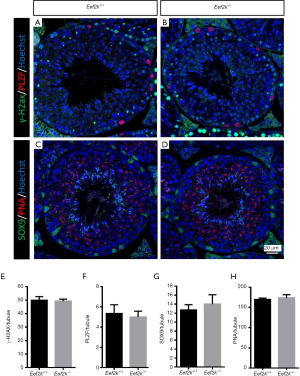
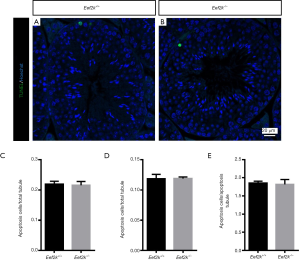
We found that male Eef2k−/− mice were fertile, with no differences in litter size relative to those associated with WT control mice (Figure 1E). Female Eef2k−/− mice exhibited normal fertility too (Figure 1E). The intercrossing of Eef2k+/− mice yielded normally-sized litters with offspring corresponding to the expected Mendelian and sex ratios (data is not shown in the text). There were also no differences in testes or epididymal tissue sizes when comparing male WT and Eef2k−/− mice (Figure 1F,G,H). Normal spermatogenic cells were observed in the PAS-stained seminiferous tubules of adult male Eef2k−/− mice, and normal quantities of spermatid cells were similarly detected in H&E-stained samples of epididymis cauda tissue from these mice (Figure 2A,B,C,D). PLZF (promyelocytic leukaemia zinc finger), γ-H2ax (phosphorylated H2A histone family member X), PNA (peanut agglutinin), and Sox9 (SRY-box 9) signals were used to identify spermatogonia, spermatocytes, spermatids, and Sertoli cells respectively (25-29), and we were able to observe all four of these cell types in testis sections from WT and Eef2k-/- mice (Figure 3A,B,C,D,E,F,G,H, Table S3). TUNEL staining analyses of testicular tissue sections also revealed comparable numbers of apoptotic cells per tubule and apoptotic tubule ratios in WT and Eef2k-/- mice (Figure 4A,B,C,D,E, Table S4).
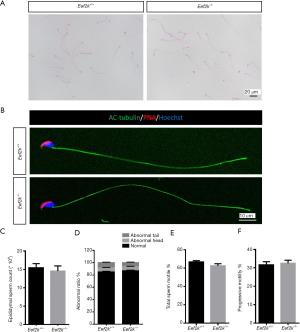
Eef2k−/− mice exhibited normal spermatozoa
No significant morphological differences were observed in the spermatozoa of male Eef2k−/− mice relative to WT mice, nor were any alterations in epididymal sperm counts detected between these two groups of mice (Figure 5A,B,C,D). Morphological analyses also revealed the ratio of normal sperm in male Eef2k
Discussion
We herein employed a CRISPR/Cas9 approach to explore the functional importance of Eef2k in spermatogenesis and male fertility by generating Eef2k-knockout mice. Our histological, immunofluorescent, and TUNEL staining assays failed to reveal any differences in spermatogenesis between Eef2k-knockout mice and WT mice, suggesting that Eef2k is dispensable in the context of normal germ cell functionality.
We had previously shown that Tcte1-knockout male mice were sterile as a consequence of asthenozoospermia despite exhibiting morphologically normal sperm (9). We found Eef2k to be significantly downregulated in the testes of these Tcte1-knockout mice in quantitative proteomics analyses (9), leading us to explore the functional impact of Eef2k knockout on spermatogenesis. We therefore hypothesized that altered Eef2k expression may impact Eef2 activity and thereby disrupt normal protein synthesis within murine sperm.
Members of the eukaryotic elongation factor protein family are important mediators of elongation during the mRNA translation (30). Eef2 is an important member of this protein family that facilitates GTP-dependent nascent protein chain translocation from the ribosomal A-site to the P-site (31), thereby regulating protein synthesis. The Eef2k kinase is responsible for phosphorylating Eef2 and thereby inhibiting its functionality, ultimately modulating translation and protein synthesis within affected cells (10,11,32).
Eef2k has been extensively studied in the context of tumorigenesis and cancer treatment (33). In tumor cells, Eef2k plays a protective role, improving the tolerance of these highly proliferative cells to energy depletion and nutrient deprivation conditions. Because they grow rapidly, cancer cells must efficiently synthesize proteins, thereby creating significant metabolic demands and requirements for substantial amino acid availability. The ability to adapt to nutrient deprivation conditions is thus essential for these cells to live when local nutrient supplies have been exhausted. Several tumor types exhibit Eef2k activation and overexpression that is associated with enhanced survival, particularly under conditions of nutrient deprivation (34-36) or acidosis (37). By disrupting translational elongation, Eef2k can effectively augment nutrient deprivation resistance (12). Through a positive feedback loop associated with Eef2k, AMPK can reduce ERK1/2 activity and the sensitivity of tumor cells to nutrition deprivation (38). Consistent with this, Eef2k overexpression was sufficient to protect RaxV12-transformed NIH3T3 cells from caloric restriction-induced cell death in vivo (12). Eef2k can additionally promote esophageal squamous cell carcinoma (ESCC) cell proliferation, migration, and invasion, and can promote the growth of ESCC tumors in xenograft model mice (16). Eef2k can also aid cell migration (14,16,39-42), potentially thereby modulating cancer progression. Spermatogonial cells are also highly proliferative in order to facilitate appropriate sperm production (43,44). As such, we hypothesized that Eef2k may similarly regulate spermatogonial cell proliferation by altering the differentiation or self-renewal of these cells. However, as we observed no differences in the morphology or motility of sperm in our Eef2k-knockout mice, our data suggest that this protein is dispensible in this context. This analysis revealed that the loss of Eef2k expression did not adversely impact sperm motility, morphology, or progressive motility in these mice.
Overall, our data indicate that Eef2k does not play a key role in murine spermatogenesis, and they provide a basis for future analyses of the mechanistic basis for spermatogenesis in these mice.
Conclusions
EEF2K is a protein that is expressed at low levels in asthenospermatic human sperm and Tcte1-knockout mouse sperm. However, the knockout of this protein did not adversely impact fertility, spermatogenesis, or development in male C57BL/6 mice. As such, these results indicate that Eef2k is dispensable in the context of male fertility. Due to the limited energy and funds, there is no further research in proteomes of the germ cells.
Acknowledgments
Funding: This work was supported by the National Key Research and Development Program of China 2016YFA0500902 (to ML); Natural Science Foundation of China (81801430 to XS, 32070842, 31771654 and 31530047 to ML); the Natural Science Foundation of Jiangsu Province (Grants No. BK20190081 to ML); and Qing Lan Project (to ML). Science development fund of Nanjing Medical University NMUB2018131 to XS.
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at http://dx.doi.org/10.21037/tau-21-18
Data Sharing Statement: available at http://dx.doi.org/10.21037/tau-21-18
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tau-21-18). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Experiments were performed under a project license (No.: IACUC-2004020) granted by the Animal Ethical and Welfare Committee, in compliance with the Institutional Animal Care and Use Committee of Nanjing Medical University guidelines.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Agarwal A, Baskaran S, Parekh N, et al. Male infertility. Lancet 2021;397:319-33. [Crossref] [PubMed]
- Lee JA, Ramasamy R. Indications for the use of human chorionic gonadotropic hormone for the management of infertility in hypogonadal men. Transl Androl Urol 2018;7:S348-52. [Crossref] [PubMed]
- Durairajanayagam D, Agarwal A, Ong C. Causes, effects and molecular mechanisms of testicular heat stress. Reprod Biomed Online 2015;30:14-27. [Crossref] [PubMed]
- Sansone A, Di Dato C, de Angelis C, et al. Smoke, alcohol and drug addiction and male fertility. Reprod Biol Endocrinol 2018;16:3. [Crossref] [PubMed]
- Krausz C, Riera-Escamilla A. Genetics of male infertility. Nat Rev Urol 2018;15:369-84. [Crossref] [PubMed]
- Signore F, Gulia C, Votino R, et al. The Role of Number of Copies, Structure, Behavior and Copy Number Variations (CNV) of the Y Chromosome in Male Infertility. Genes (Basel) 2019;11:40. [Crossref] [PubMed]
- Zhang Y, Guo R, Cui Y, et al. An essential role for PNLDC1 in piRNA 3’ end trimming and male fertility in mice. Cell Res 2017;27:1392-6. [Crossref] [PubMed]
- Babakhanzadeh E, Nazari M, Ghasemifar S, et al. Some of the Factors Involved in Male Infertility: A Prospective Review. Int J Gen Med 2020;13:29-41. [Crossref] [PubMed]
- Castaneda JM, Hua R, Miyata H, et al. TCTE1 is a conserved component of the dynein regulatory complex and is required for motility and metabolism in mouse spermatozoa. Proc Natl Acad Sci U S A 2017;114:E5370-8. [Crossref] [PubMed]
- Kenney JW, Moore CE, Wang X, et al. Eukaryotic elongation factor 2 kinase, an unusual enzyme with multiple roles. Adv Biol Regul 2014;55:15-27. [Crossref] [PubMed]
- Wang X, Xie J, Proud CG. Eukaryotic elongation factor 2 kinase (eEF2K) in cancer. Cancers 2017;9:162. [Crossref] [PubMed]
- Leprivier G, Remke M, Rotblat B, et al. The eEF2 kinase confers resistance to nutrient deprivation by blocking translation elongation. Cell 2013;153:1064-79. [Crossref] [PubMed]
- Meric-Bernstam F, Chen H, Akcakanat A, et al. Aberrations in translational regulation are associated with poor prognosis in hormone receptor-positive breast cancer. Breast Cancer Res 2012;14:R138. [Crossref] [PubMed]
- Tekedereli I, Alpay SN, Tavares CD, et al. Targeted silencing of elongation factor 2 kinase suppresses growth and sensitizes tumors to doxorubicin in an orthotopic model of breast cancer. PLoS One 2012;7:e41171 [Crossref] [PubMed]
- Zhang Y, Cheng Y, Zhang L, et al. Inhibition of eEF-2 kinase sensitizes human glioma cells to TRAIL and down-regulates Bcl-xL expression. Biochem Biophys Res Commun 2011;414:129-34. [Crossref] [PubMed]
- Zhu H, Song H, Chen G, et al. eEF2K promotes progression and radioresistance of esophageal squamous cell carcinoma. Radiother Oncol 2017;124:439-47. [Crossref] [PubMed]
- Wang X, Xie W, Yao Y, et al. The heat shock protein family gene Hspa1l in male mice is dispensable for fertility. PeerJ 2020;8:e8702 [Crossref] [PubMed]
- Zhang J, Zhang X, Zhang Y, et al. Normal spermatogenesis in Fank1 (fibronectin type 3 and ankyrin repeat domains 1) mutant mice. PeerJ 2019;7:e6827 [Crossref] [PubMed]
- Mahmood T, Yang P-C. Western blot: technique, theory, and trouble shooting. North American journal of medical sciences 2012;4:429. [Crossref] [PubMed]
- Ahmed EA, de Rooij DG. Staging of mouse seminiferous tubule cross-sections. Meiosis: Springer, 2009:263-77.
- Clermont Y, Leblond C. Spermiogenesis of man, monkey, ram and other mammals as shown by the “periodic acid-Schiff” technique. Am J Anat 1955;96:229-53. [Crossref] [PubMed]
- Crowley LC, Marfell BJ, Waterhouse NJ. Detection of DNA fragmentation in apoptotic cells by TUNEL. Cold Spring Harbor Protocols 2016;2016:pdb. prot087221.
- Kyrylkova K, Kyryachenko S, Leid M, et al. Detection of apoptosis by TUNEL assay. Odontogenesis. Springer, 2012:41-7.
- Amann RP, Waberski D. Computer-assisted sperm analysis (CASA): capabilities and potential developments. Theriogenology 2014;81:5-17.e1. [Crossref] [PubMed]
- Buaas FW, Kirsh AL, Sharma M, et al. Plzf is required in adult male germ cells for stem cell self-renewal. Nat Genetics 2004;36:647-52. [Crossref] [PubMed]
- Costoya JA, Hobbs RM, Barna M, et al. Essential role of Plzf in maintenance of spermatogonial stem cells. Nat Genetics 2004;36:653-9. [Crossref] [PubMed]
- Fernandez-Capetillo O, Mahadevaiah SK, Celeste A, et al. H2AX is required for chromatin remodeling and inactivation of sex chromosomes in male mouse meiosis. Developmental Cell 2003;4:497-508. [Crossref] [PubMed]
- Nakata H, Wakayama T, Asano T, et al. Identification of sperm equatorial segment protein 1 in the acrosome as the primary binding target of peanut agglutinin (PNA) in the mouse testis. Histochem Cell Biol 2017;147:27-38. [Crossref] [PubMed]
- Daigle M, Roumaud P, Martin LJ. Expressions of Sox9, Sox5, and Sox13 transcription factors in mice testis during postnatal development. Mol Cell Biochem 2015;407:209-21. [Crossref] [PubMed]
- Browne GJ, Proud CG. Regulation of peptide-chain elongation in mammalian cells. Eur J Biochem 2002;269:5360-8. [Crossref] [PubMed]
- Susorov D, Zakharov N, Shuvalova E, et al. Eukaryotic translation elongation factor 2 (eEF2) catalyzes reverse translocation of the eukaryotic ribosome. J Biol Chem 2018;293:5220-9. [Crossref] [PubMed]
- Kaul G, Pattan G, Rafeequi T. Eukaryotic elongation factor-2 (eEF2): its regulation and peptide chain elongation. Cell Biochem Funct 2011;29:227-34. [Crossref] [PubMed]
- De Gassart A, Demaria O, Panes R, et al. Pharmacological eEF2K activation promotes cell death and inhibits cancer progression. EMBO Rep 2016;17:1471-84. [Crossref] [PubMed]
- Leprivier G, Rotblat B, Khan D, et al. Stress-mediated translational control in cancer cells. Biochim Biophys Acta 2015;1849:845-60. [Crossref] [PubMed]
- Leprivier G, Sorensen PH. How does oncogene transformation render tumor cells hypersensitive to nutrient deprivation? Bioessays 2014;36:1082-90. [Crossref] [PubMed]
- Manning BD. Adaptation to starvation: translating a matter of life or death. Cancer Cell 2013;23:713-5. [Crossref] [PubMed]
- Xie J, Mikolajek H, Pigott CR, et al. Molecular Mechanism for the Control of Eukaryotic Elongation Factor 2 Kinase by pH: Role in Cancer Cell Survival. Mol Cell Biol 2015;35:1805-24. [Crossref] [PubMed]
- Tong S, Zhou T, Meng Y, et al. AMPK decreases ERK1/2 activity and cancer cell sensitivity to nutrition deprivation by mediating a positive feedback loop involving eEF2K. Oncol Lett 2020;20:61-6. [Crossref] [PubMed]
- Bayraktar R, Pichler M, Kanlikilicer P, et al. MicroRNA 603 acts as a tumor suppressor and inhibits triple-negative breast cancer tumorigenesis by targeting elongation factor 2 kinase. Oncotarget 2017;8:11641. [Crossref] [PubMed]
- Hamurcu Z, Ashour A, Kahraman N, et al. FOXM1 regulates expression of eukaryotic elongation factor 2 kinase and promotes proliferation, invasion and tumorgenesis of human triple negative breast cancer cells. Oncotarget 2016;7:16619. [Crossref] [PubMed]
- Shi Q, Xu X, Liu Q, et al. MicroRNA-877 acts as a tumor suppressor by directly targeting eEF2K in renal cell carcinoma. Oncol Lett 2016;11:1474-80. [Crossref] [PubMed]
- Usui T, Nijima R, Sakatsume T, et al. Eukaryotic elongation factor 2 kinase controls proliferation and migration of vascular smooth muscle cells. Acta Physiol (Oxf) 2015;213:472-80. [Crossref] [PubMed]
- Mäkelä JA, Hobbs RM. Molecular regulation of spermatogonial stem cell renewal and differentiation. Reproduction 2019;158:R169-87. [Crossref] [PubMed]
- Chen SR, Liu YX. Regulation of spermatogonial stem cell self-renewal and spermatocyte meiosis by Sertoli cell signaling. Reproduction 2015;149:R159-67. [Crossref] [PubMed]


