
Development and validation of the prognostic value of the immune-related genes in clear cell renal cell carcinoma
Introduction
Clear cell renal cell carcinoma (ccRCC) representing approximately 75% of RCC cases with more than 175000 deaths per year (1). Although surgical resection is effective for localized RCC, about one-third cases suffered recurrences and metastases with worse prognosis (2). Various molecular signatures of ccRCC implies that distinct survival advantages exist in the certain subtypes (3,4). Owing to heterogeneity, discovering reliable molecular biomarkers can help to improve prognostic determination and guide clinical decision. Actually, RCC is believed to be an immunogenic tumor for long time. Interleukin 2 (IL-2) and interferon alpha (IFN-a) were used for therapeutic regimens for advanced RCC in the 1990s to early 2000s, and the incidence of complete remission was about 3–5% (5,6). Recently, trail of KEYNOTE-426 and JAVELIN Renal 101 demonstrates using PD-1 immune checkpoint inhibitor-based combination regimens as the first line setting can significantly improve advanced RCC survival, which have been approved by FDA (7,8). Therefore, further exploration of immune related molecular network in RCC definitely helps to develop comprehensive understanding of immune evasion and provide insights into making therapeutic strategy. Immune molecular regulation is the key mechanism for host innate immunity and immune surveillance. It is necessary to explore clinical significance of immune-related biomarkers, especially immune-related genes (IRGs) which could predict prognosis of patients, and potentially portrait tumor microenvironment (TME) (9,10).
In this study, we identified 4 immune-related genes (CRABP2, LTB4R, PTGER1 and TEK) through integrated analyses of mRNA expression data from TCGA database and independently assessed. Multivariate Cox proportional hazards models were constructed by 4 genes and validated the accuracy in an external ICGC dataset. Moreover, we investigated a high proportion of CD8 T cell, regulatory T cell and NK cell in the unfavorable-risk group. High levels of immune response suppressors (PD-1, CTLA4, TNFRSF9, TIGIT and LAG3) were observed in unfavorable-risk group and positively correlated with risk score. According to aforementioned data, a nomogram was well established for clinical use and also externally validated its superior power by ICGC data.
We present the following article in accordance with the TRIPOD reporting checklist (available at http://dx.doi.org/10.21037/tau-20-1348).
Methods
Study design and dataset information
The work flow of our study is shown in Figure 1. The expression profile and clinical data of 539 ccRCC patients in TCGA-KIRC dataset were downloaded from TCGA portal (online URL: https://cdn.amegroups.cn/static/public/tau-20-1348-1.pdf) and 91 RCC patients from ICGC database were downloaded from ICGC portal (online URL: https://dcc.icgc.org/). Immune-related genes list was downloaded from the ImmPort database (online URL: https://www.immport.org/home/). All data were preprocessed in R software (online URL: https://www.r-project.org/; version 3.6.0;). 518 patients in TCGA-KIRC cohort and 91 patients in ICGC cohort with clinical information (Table S1 and S2) were screened for subsequent analyses. All procedures performed in this study were in accordance with the Declaration of Helsinki (as revised in 2013).
Identification of IRDEGs in TCGA-KIRC dataset
Differential analysis was conducted in TCGA-KIRC dataset through limma package (11,12), with the following cut-off: adjusted P value <0.05 and absolute log2FC >1.5. The differentially expressed genes list and immune-related genes list from the ImmPort database were uploaded into the Venn diagram online software (online URL: http://bioinformatics.psb.ugent.be/webtools/Venn/) to obtain the IRDEGs. The heatmap of IRDEGs expression was performed by pheatmap package.
Construction and validation of the risk model
518 patients in TCGA-KIRC dataset were included as a training set while 91 patients in the ICGC database were assigned as a validation set. Univariate Cox proportional hazards regression analysis was applied to identify the significant prognostic factors associated with OS, and Lasso regression was used to exclude overfitting genes. The candidate genes were analyzed in a multivariate Cox proportional hazards regression analysis to estimate their relative contributions to survival prediction. Subsequently, a prognostic model was constructed: risk score = expression of gene1 × β1 + expression of gene2 × β2 +…… + expression of genen × βn (13,14). According to the median risk score, patients were divided into two groups (favorable-risk group and unfavorable-risk group), and we applied the Kaplan-Meier and log-rank methods to test whether the survival distribution of different groups was equal. Receiver operating characteristic (ROC) curves were used to assess the predictive value of the risk model according to the areas under the respective ROC curves (AUCs). Time-dependent ROC curve analysis was conducted by using the survival ROC package (15).
RNA extraction and qRT-PCR analysis
Total RNA of 35 pairs of ccRCC and normal tissues RNA were extracted using a Trizol reagent, and 500 ng of RNA was used to synthesize cDNA, and qRT-PCR was performed on ABI system. The primer sequences are listed in Table S3.
Estimation of TME infiltration between groups
CIBERSORT, a deconvolution algorithm to characterize different cell compositions of the samples based on the immune gene signature sets, including 547 genes and 22 immune cell subtypes (16). We downloaded the result of 518 patients in TCGA cohort calculated by CIBERSORT algorithm from TIMER 2.0(Online URL: http://timer.cistrome.org/) (17) to estimate the infiltration of 22 different immune cell subtypes in the TME for further investigation of the composition and difference between favorable-risk and unfavorable-risk group. Each sample had been calculated a proportion in each cell subtype to estimate the relative abundance of TME immune infiltrating cells. Wilcox test was applied to compare the infiltration proportion of the 22 cell types between the unfavorable-risk group and the favorable-risk group.
Construction and validation of the clinical nomogram
We constructed a nomogram, which was widely used to predict the survival probability of patients in clinical (18), with the incorporation of age, gender, TNM stage, Fuhrman grade, necrosis and risk score through R rms package. We also used ROC curves to evaluate the predictive performance of nomogram at 1-, 3- and 5-year. In addition, calibration curves were used to evaluate the accuracy of the predicted survival time for 3- and 5-year OS, and decision curve analysis (DCA) was performed to evaluate the clinical application benefit between different variables.
Statistical analysis
All statistical analyses were performed by R software with the cut-off of P<0.05. Univariate and multivariate Cox proportional hazards regression analyses were used for identifying of prognosis-related IRDEGs and independent prognostic factors. Spearman correlation test was applied to analyze the correlation between the risk score and the expression of immune checkpoint genes. Survival data were calculated using the Kaplan-Meier method and the log-rank test. The relative expression level of four IRDEGs was analyzed by paired t test.
Results
Identification of IRDEGs in ccRCC
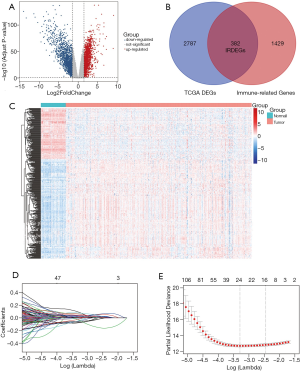
After differential analysis in TCGA-KIRC cohort, 3,169 DEGs were detected (adjusted P value <0.05 and absolute log2FC >1.5), among which 1,635 genes were upregulated, and 1534 genes were downregulated (Figure 2A). Taking the intersection of DEGs and the immune-related genes list, 382 IRDEGs were identified (Figure 2B), with 253 IRDEGs upregulated and 129 IRDEGs downregulated. The heatmap to visualize the expression of 382 IRDEGs in normal samples and tumor samples is shown in Figure 2C, and the result of differential analysis are shown in https://cdn.amegroups.cn/static/public/tau-20-1348-2.xlsx and Table S4.
Construction of prognostic model in TCGA cohort
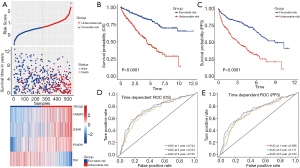
175 IRDEGs was calculated to be significantly associated with OS after univariate Cox regression analysis (P<0.05) (Table S5). Lasso regression was used to filter genes to obtain 8 candidate genes (Figure 2D,2E), which were subsequently included in the multivariate Cox regression analysis. A prognostic gene signature consisting of 4 genes was ultimately constructed with the P value <0.05 in multivariate Cox regression analysis (Table 1 and Table S6). Among these 4 genes, TEK was identified as a protective gene because of its hazard ratios (HR) value of <1, while CRABP2, LTB4R and PTGER1 were considered to be predictive genes of poor prognosis. Based on the analysis result, we constructed a computational formula: risk score = (0.074× expression level of CRABP2) + (0.165× expression level of LTB4R) + (0.052× expression level of PTGER1) + (-0.203× expression level of TEK), and the expression level was obtained by the log2-transformed FPKM+1 of each gene. Subsequently, a total of 518 patients in TCGA cohort were divided into two groups (unfavorable-risk group and favorable-risk group) according to the median risk score. Figure 3A shows the distribution of risk scores, patient survival status and the four gene expression levels in the 518 patients, which were sorted by the risk score of the four-gene signature. As the risk score increasing, the expression of the 4 IRDEGs also changed accordingly, and the prognosis of patients also became worse. Besides, as Figure 3B and 3C show, there were obvious differences in both OS and PFS between the two groups (P<0.0001). On the other hand, a time-dependent ROC was used to assess the prognostic value of the four-gene signature in the training set. The AUCs of the signature were respectively 0.744, 0.734, and 0.753 for the 1-, 3- and 5-year OS (Figure 3D) while for the 1-, 3- and 5-year PFS were 0.696, 0.711, and 0.734(Figure 3E), indicating our risk model had a good performance on predicting prognosis. Besides, further analysis showed risk score was an independent prognostic factor (HR: 3.137; 95% CI: 2.383-4.131; P<0.001) (Table 1).

Full table
External validation of prognostic model in ICGC cohort
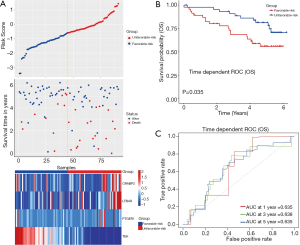
We used the data from ICGC database as an external validation (Figure 4). The distribution of risk scores and survival status of patients, as well as the expression level of the 4 IRDEGs in ICGC cohort, were shown in Figure 4A and it was observed that the expression had an obvious difference between unfavorable-risk group and favorable-risk group. Besides, Kaplan-Meier analysis indicated that unfavorable-risk group was significantly associated with a poor prognosis (P<0.05), consistently with the above results (Figure 4B). And the AUCs of ROC analysis were respectively 0.635, 0.638 and 0.635 at 1-, 3- and 5-year OS value, indicating the stability of risk model in different cohorts (Figure 4C).
Validation in clinical samples
We used 35 pairs of ccRCC (Table S7) and normal tissues to detect the expression of the four IRDEGs. The results showed LTB4R was high expressed in ccRCC compared with normal tissues, and CRABP2, PTGER1, TEK were low expressed, consistent with the expression data of TCGA database (Figure 5).
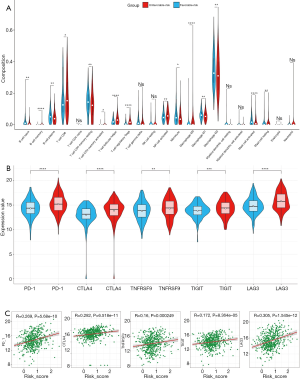
TME immune cell infiltration analysis and immune checkpoints analysis
We summarized the result of 518 ccRCC patients calculated by CIBERSORT algorithm (Figure S1) and compared all the immune cell subtypes in two groups (Table S8). Infiltration proportion of partial cell subtypes have an obvious difference between two groups, among which mainly CD8 T cell, follicular helper T cell, regulatory T cell, activated NK cell, M0 Macrophage have a higher infiltration proportion in the unfavorable-risk group, while M1 Macrophage, M2 Macrophage and other cell types have a lower proportion (Figure 6A). We further explored the expression of the T cell exhaustion-related markers and immunomodulators (PD-1, CTLA4, TNFRSF9, TIGIT, LAG3) in two groups and found all markers in the unfavorable-risk group were upregulated, indicating an immunosuppressive and exhausted phenotype in the unfavorable-risk group (Figure 6B). Subsequent correlation analysis also showed a positive correlation between risk score and the above markers (Figure 6C). Based on the above analyses, we found two groups had a significant distinct pattern of immune infiltration, which may lead to different survival benefits.
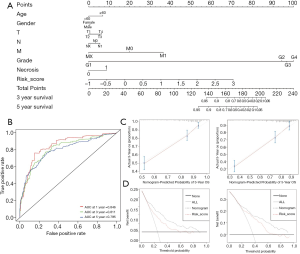
Construction and validation of the nomogram
We constructed a nomogram containing age, gender, TNM stage, Fuhrman grade, necrosis and risk score to predict 3- and 5-year survival probability. Each variable had a corresponding score (Table S9), and an overall score could be finally calculated to predict the survival probability at the corresponding time (Figure 7A). To validate the performance of nomogram, we conducted the ROC analysis, and the result showed respective AUCs were 0.811 and 0.795 in the TCGA cohort (Figure 7B). The calibration curves showed good consistency between the actual and predicted outcomes of 3- and 5-year OS (Figure 7C). Decision curve analysis (DCA) was also conducted, and all variable curves were above the two solid curves. The curve of the nomogram was above the curve of risk score at 3- and 5-year (Figure 7D), indicating nomogram had a better clinical net benefit. We also constructed another nomogram containing age, gender, TNM stage, and risk score. And the AUCs were 0.811, 0.786 while in the validation set were 0.728, 0.713 at 3-, 5-year, suggesting its stability and effectiveness (Figures S2, S3). Calibration curves and DCA also showed the robustness of our nomogram.
Discussion
Immune related genes (IRGs) play an important role in tumor immune infiltration as well as tumor progression in ccRCC (19,20) and strongly influence complicate soluble factors secretion, which correlate with therapeutic response and clinical outcome (21). IRGs based prognostic model have been successfully developed for hepatocellular, colorectal, lung and bladder cancers (22-25). In this study, we screened and validated CRABP2, LTB4R, PTGER1 and TEK from TCGA database as potent IRGs to predict the survival risk in ccRCC patients. Multivariate Cox proportional hazards models were constructed to stratify patients based on the 4 genes signature. Laboratory q-PCR confirmed LTB4R upregulated and CRABP2, PTGER1, TEK downregulated in 35 pairs of ccRCC and normal tissues.
LTB4R is a receptor of Leukotriene B4 and is expressed mainly in leukocytes like granulocytes, macrophages and eosinophils (26). Several studies implied it was involved in CD8 T cells recruiting (27,28). The neutrophilic influx induced by LTB4 increases the pro-tumorigenic activity of tumor-associated neutrophils through releasing reactive oxygen species, inflammatory cytokines and injuring innate immune response (29,30). CRABP2 was responding in retinoic acid (RA) transduction as a tumor suppressive pathway (31). However, artificially overexpressing CRABP2 in Caki-2 cells did not exhibit a significant change in RA sensitivity. Our data indicated CRABP2 was lowly expressed in ccRCC samples, which was consistent with previous study (32). Although the exactly role of CRABP2 in RCC is not clear yet, our data showed high CARBP2 expression was an independent predictor factor for worse prognosis. Further investigations are necessary to define other molecules involved in CARBP2 mediated RA signaling and metabolism in RCC. PTGER1 is one of the receptors of prostaglandin and it couples with G-proteins to activate protein kinase C (33). Previous study implied blocking PTGER1 could suppress immunosuppressive function of Treg and subsequently inhibit tumor growth in colon cancer (34). In ccRCC, our result also exhibited high PTGER1 expression was correlated with worse prognosis. TEK encodes Tie2, which cooperate with VEGFs as critical regulators of vascular development (35). Actually, mechanisms of angiogenesis are extremely complex depending on different tumor stage and content, therefore, the prediction role of Tie2 is inconsistent among different tumor types (36). Increasing Tie2 expression correlated with high metastasis risk and poor survival among breast cancer and glioblastoma patients (37,38). However, in ccRCC, our data implied downregulation of TEK associated with a poor prognosis which was also demonstrated in previous studies (39,40). Low expression of TEK has been noted in aggressive ccRCC for years (41). Recently, when compared gene prevalence between nonmetastatic and metastatic ccRCC by target next generation sequencing from 106 sporadic cases, higher frequencies of TEK mutations involved in metastatic cohort (42). Since Tie2 signaling influences vascular permeability, low expression of Tie2 may potentiate inflammatory cells migration into tumor microenvironment (43). Inflammatory cytokines such as TNF-α, IL-6, CXCL8 induces a more aggressive tumor phenotype via immune surveillance and form premetastatic niche. Overall, we assume TEK is a tumor suppressor in ccRCC, further studies are needed in future.
Solid tumors usually disrupt tumor target immune response by subvert immune surveillance. Tumor immune signature is highly correlated with tumor prognosis and response to immunotherapy. When using a 34-gene expression signature, ccRCC can be characterized into high angiogenesis tumor with improved prognosis or high immunocytes tumor with worse survival (44). Tumor infiltrating lymphocytes in ccRCC were analyzed by gene expression and cytometry phenotyping, the result implied more poorly cytotoxic CD8 T cell, Treg infiltrate in unfavorable risk group (45). In this current study, we also investigated high proportion of CD8 T cell, Treg, NK cell in unfavorable risk group through CIBERSORT algorithm, which indicated that our novel prediction model could properly distinct patient into different immunological features. Besides, a group of immunomodulators (PD-1, CTLA4, TNFRSF9, TIGIT and LAG3) was significantly correlated with our risk score, and confirmed our risk model stably stratified patients from immune evasion perspective.
In order to increase prediction accuracy, we developed a clinical nomogram with age, gender, TNM stage, Fuhrman grade, necrosis and risk score. This nomogram obtained an AUC of 0.846, 0.811 and 0.795 in predicting the possibility of survival at 1-, 3- and 5-year respectively. As there is lack of Fuhrman grade, necrosis information in ICGC database, we removed these two factors from original nomogram for validation. The modified nomogram consistently obtained a relatively high AUC of 0.755, 0.728 and 0.713 in survival prediction at 1-, 3- and 5-year separately in ICGC data. Hence, based on the 4 immune related genes CRABP2, LTB4R, PTGER1 and TEK, we successfully constructed a prognostic risk model for ccRCC and externally validated its accuracy. Defective T-cells and aberrant expression suppressive immunomodulators lead tumor be more aggressive. Owing to data we obtained from public database, further independent validation in prospective studies is needed.
Acknowledgments
English Language Editor: Dr. M. Mumin.
Funding: Our work was supported by National Natural Science Foundation of China (No. 81202011, 81572522); Science and Technology Planning Project of Guangdong Province, China (No. 2016A020215235); Pearl River Nova Program of Guangzhou, China (No. 201710010057).
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at http://dx.doi.org/10.21037/tau-20-1348
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tau-20-1348
Peer Review File: Available at http://dx.doi.org/10.21037/tau-20-1348
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tau-20-1348). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in this study were in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Gupta K, Miller JD, Li JZ, et al. Epidemiologic and socioeconomic burden of metastatic renal cell carcinoma (mRCC): a literature review. Cancer Treat Rev 2008;34:193-205. [Crossref] [PubMed]
- Rini B, Goddard A, Knezevic D, et al. A 16-gene assay to predict recurrence after surgery in localised renal cell carcinoma: development and validation studies. Lancet Oncol 2015;16:676-85. [Crossref] [PubMed]
- Brooks SA, Brannon AR, Parker JS, et al. ClearCode34: A prognostic risk predictor for localized clear cell renal cell carcinoma. Eur Urol 2014;66:77-84. [Crossref] [PubMed]
- Bukowski RM. Natural history and therapy of metastatic renal cell carcinoma: the role of interleukin-2. Cancer 1997;80:1198-220. [Crossref] [PubMed]
- Motzer RJ, Bacik J, Murphy BA, et al. Interferon-alfa as a comparative treatment for clinical trials of new therapies against advanced renal cell carcinoma. J Clin Oncol 2002;20:289-96. [Crossref] [PubMed]
- Rini BI, Plimack ER, Stus V, et al. Pembrolizumab plus Axitinib versus Sunitinib for Advanced Renal-Cell Carcinoma. N Engl J Med 2019;380:1116-27. [Crossref] [PubMed]
- Motzer RJ, Penkov K, Haanen J, et al. Avelumab plus Axitinib versus Sunitinib for Advanced Renal-Cell Carcinoma. N Engl J Med 2019;380:1103-15. [Crossref] [PubMed]
- Chen YP, Wang YQ, Lv JW, et al. Identification and validation of novel microenvironment-based immune molecular subgroups of head and neck squamous cell carcinoma: implications for immunotherapy. Ann Oncol 2019;30:68-75. [Crossref] [PubMed]
- Ji RR, Chasalow SD, Wang L, et al. An immune-active tumor microenvironment favors clinical response to ipilimumab. Cancer Immunol Immunother 2012;61:1019-31. [Crossref] [PubMed]
- Phipson B, Lee S, Majewski IJ, et al. Robust Hyperparameter Estimation Protects against Hypervariable Genes and Improves Power to Detect Differential Expression. Ann Appl Stat 2016;10:946-63. [Crossref] [PubMed]
- Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43:e47 [Crossref] [PubMed]
- Huang R, Liao X, Li Q. Identification and validation of potential prognostic gene biomarkers for predicting survival in patients with acute myeloid leukemia. Onco Targets Ther 2017;10:5243-54. [Crossref] [PubMed]
- Zhou M, Zhao H, Wang Z, et al. Identification and validation of potential prognostic lncRNA biomarkers for predicting survival in patients with multiple myeloma. J Exp Clin Cancer Res 2015;34:102. [Crossref] [PubMed]
- Heagerty PJ, Zheng Y. Survival model predictive accuracy and ROC curves. Biometrics 2005;61:92-105. [Crossref] [PubMed]
- Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods 2015;12:453-7. [Crossref] [PubMed]
- Li T, Fan J, Wang B, et al. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res 2017;77:e108-10. [Crossref] [PubMed]
- Balachandran VP, Gonen M, Smith JJ, et al. Nomograms in oncology: more than meets the eye. Lancet Oncol 2015;16:e173-80. [Crossref] [PubMed]
- Xu WH, Shi SN, Xu Y, et al. Prognostic implications of Aquaporin 9 expression in clear cell renal cell carcinoma. J Transl Med 2019;17:363. [Crossref] [PubMed]
- Du GW, Yan X, Chen Z, et al. Identification of transforming growth factor beta induced (TGFBI) as an immune-related prognostic factor in clear cell renal cell carcinoma (ccRCC). Aging (Albany NY) 2020;12:8484-505. [Crossref] [PubMed]
- Wu T, Dai Y. Tumor microenvironment and therapeutic response. Cancer Lett 2017;387:61-8. [Crossref] [PubMed]
- Luo C, Lei M, Zhang Y, et al. Systematic construction and validation of an immune prognostic model for lung adenocarcinoma. J Cell Mol Med 2020;24:1233-44. [Crossref] [PubMed]
- Qiu H, Hu X, He C, et al. Identification and Validation of an Individualized Prognostic Signature of Bladder Cancer Based on Seven Immune Related Genes. Front Genet 2020;11:12. [Crossref] [PubMed]
- Wang Z, Zhu J, Liu Y, et al. Development and validation of a novel immune-related prognostic model in hepatocellular carcinoma. J Transl Med 2020;18:67. [Crossref] [PubMed]
- Zhao X, Liu J, Liu S, et al. Construction and Validation of an Immune-Related Prognostic Model Based on TP53 Status in Colorectal Cancer. Cancers (Basel) 2019;11:1722. [Crossref] [PubMed]
- Tager AM, Luster AD. BLT1 and BLT2: the leukotriene B(4) receptors. Prostaglandins Leukot Essent Fatty Acids 2003;69:123-34. [Crossref] [PubMed]
- Chheda ZS, Sharma RK, Jala VR, et al. Chemoattractant Receptors BLT1 and CXCR3 Regulate Antitumor Immunity by Facilitating CD8+ T Cell Migration into Tumors. J Immunol 2016;197:2016-26. [Crossref] [PubMed]
- Sharma RK, Chheda Z, Jala VR, et al. Expression of leukotriene B(4) receptor-1 on CD8(+) T cells is required for their migration into tumors to elicit effective antitumor immunity. J Immunol 2013;191:3462-70. [Crossref] [PubMed]
- Galdiero MR, Garlanda C, Jaillon S, et al. Tumor associated macrophages and neutrophils in tumor progression. J Cell Physiol 2013;228:1404-12. [Crossref] [PubMed]
- Fridlender ZG, Sun J, Kim S, et al. Polarization of tumor-associated neutrophil phenotype by TGF-beta: "N1" versus "N2" TAN. Cancer Cell 2009;16:183-94. [Crossref] [PubMed]
- Fu YS, Wang Q, Ma JX, et al. CRABP-II methylation: a critical determinant of retinoic acid resistance of medulloblastoma cells. Mol Oncol 2012;6:48-61. [Crossref] [PubMed]
- Goelden U, Pfoertner S, Hansen W, et al. Expression and functional influence of cellular retinoic acid-binding protein II in renal cell carcinoma. Urol Int 2005;75:269-76. [Crossref] [PubMed]
- Sugimoto Y, Narumiya S. Prostaglandin E receptors. J Biol Chem 2007;282:11613-7. [Crossref] [PubMed]
- O'Callaghan G, Ryan A, Neary P, et al. Targeting the EP1 receptor reduces Fas ligand expression and increases the antitumor immune response in an in vivo model of colon cancer. Int J Cancer 2013;133:825-34. [Crossref] [PubMed]
- Ramsauer M, D'Amore PA. Getting Tie(2)d up in angiogenesis. J Clin Invest 2002;110:1615-7. [Crossref] [PubMed]
- Sharma S, Sharma MC, Sarkar C. Morphology of angiogenesis in human cancer: a conceptual overview, histoprognostic perspective and significance of neoangiogenesis. Histopathology 2005;46:481-9. [Crossref] [PubMed]
- Dales JP, Garcia S, Bonnier P, et al. Tie2/Tek expression in breast carcinoma: correlations of immunohistochemical assays and long-term follow-up in a series of 909 patients. Int J Oncol 2003;22:391-7. [Crossref] [PubMed]
- Zheng S, Tao W. Identification of Novel Transcriptome Signature as a Potential Prognostic Biomarker for Anti-Angiogenic Therapy in Glioblastoma Multiforme. Cancers (Basel) 2020;12:2368. [PubMed]
- Shen C, Liu J, Wang J, et al. Development and validation of a prognostic immune-associated gene signature in clear cell renal cell carcinoma. Int Immunopharmacol 2020;81:106274 [Crossref] [PubMed]
- Ha M, Son YR, Kim J, et al. TEK is a novel prognostic marker for clear cell renal cell carcinoma. Eur Rev Med Pharmacol Sci 2019;23:1451-8. [PubMed]
- Kosari F, Parker AS, Kube DM, et al. Clear cell renal cell carcinoma: gene expression analyses identify a potential signature for tumor aggressiveness. Clin Cancer Res 2005;11:5128-39. [Crossref] [PubMed]
- Meng H, Jiang X, Cui J, et al. Genomic Analysis Reveals Novel Specific Metastatic Mutations in Chinese Clear Cell Renal Cell Carcinoma. Biomed Res Int 2020;2020:2495157 [Crossref] [PubMed]
- Parikh SM. Angiopoietins and Tie2 in vascular inflammation. Curr Opin Hematol 2017;24:432-8. [Crossref] [PubMed]
- Iglesia MD, Parker JS, Hoadley KA, et al. Genomic Analysis of Immune Cell Infiltrates Across 11 Tumor Types. J Natl Cancer Inst 2016;108:djw144 [Crossref] [PubMed]
- Giraldo NA, Becht E, Vano Y, et al. Tumor-Infiltrating and Peripheral Blood T-cell Immunophenotypes Predict Early Relapse in Localized Clear Cell Renal Cell Carcinoma. Clin Cancer Res 2017;23:4416-28. [Crossref] [PubMed]



