
This article has an erratum available at: http://dx.doi.org/10.21037/tau-2022-04 the article has been update on 2022-10-18 at here.
A partial loss-of-function variant in GNRNR gene in a Chinese cohort with idiopathic hypogonadotropic hypogonadism
Introduction
Patients with idiopathic hypogonadotropic hypogonadism (IHH) present with incomplete puberty development, low gonadotropin levels, and infertility. Most patients are diagnosed in their late adolescence or in early adulthood. IHH is a rare genetically heterogeneous disorder caused by the deficient development and migration of gonadotropin-releasing hormone (GnRH) neurons and deficient GnRH secretion and action (1). The impaired hypothalamic-pituitary-gonadal axis further leads to steroidogenic and gametogenic disorders. The prevalence of IHH is approximately 1:10,000 in males and 1:50,000 in females (2). IHH has high phenotypic and genetic heterogeneity. IHH with olfactory disorder (50–60% of IHH) is known as Kallmann syndrome (KS), while IHH with a normal sense of smell is described as normosmic IHH (nIHH) (3). There are multiple inheritance mode of causal genes, including autosomal dominant, autosomal recessive, X-linked recessive, as well as digenic and oligogenic transmission (4).
To date, approximately >30 genes has been discovered to be implicated in IHH, including ANOS1, GNRH1/GNRHR, FGF8/FGFR1, PROK2/PROKR2, SEMA3A/PLXNA1, KISS1/KISS1R, SEMA3E, NSMF, HS6ST1, WDR11, SOX10, etc. (5). GNRHR (GnRH receptor) is the first gene identified as a monogenic cause of nIHH (6). GNRHR is located in chromosome 4q21.2, and encodes a seven-transmembrane G protein coupled receptor (7). GNRHR and its ligand, GnRH, play a crucial role in the secretion of anterior pituitary gonadotropes (8). Their binding facilitates an increase in inositol trisphosphate and calcium mobilization, and ultimately triggers the synthesis and secretion of the follicle-stimulating and luteinizing hormones (9). So far, at least fifty-five variants in the GNRHR gene have been found among nIHH patients, and the homozygous and compound heterozygous mutations are very common (10). However, it is noteworthy that heterozygous GNRHR mutations were often identified among IHH patients (11,12). A genetic study on a large IHH cohort showed that frequency of heterozygous GNRHR RSVs reached 2.5% (22/863), significantly higher than the healthy control population (12). An recent study also revealed that this heterozygous RSV was found in the three sporadic IHH patients, without carrying other common IHH gene mutations (11). Therefore, researchers speculated that monoallelic GNRHR RSVs are associated with IHH, but there was limited literature regarding their functional implications.
The identification of the more GNRHR RSVs is not only beneficial to the early genetic diagnosis of IHH, but also conducive to the development of individualized treatment for patients. For example, if the IHH patients with pathogenic GNRHR RSVs receive pulsatile GnRH treatment, it is predicted that their therapeutic response would not be very good. Gonadotropin treatment will help these patients achieve better therapeutic results. In addition, GnRH agonists (GnRH-a) are widely used in the treatment of prostate cancer, endometriosis and uterus adenomyosis. It is reasonable to speculate that the GNRHR RSVs have a negative effect on the response to GnRH-a in these diseases. The aim of this study was to investigate GNRHR mutation profile in a Chinese IHH cohort, and to characterize the clinical manifestation and therapeutic outcome. In vitro functional analysis of GNRHR rare sequencing variants (RSVs) was also performed. We present the following article in accordance with the MDAR checklist (available at http://dx.doi.org/10.21037/tau-20-1390).
Methods
Patients
From 2005 to 2013, a total of 153 male patients were recruited at the Urology Clinic of Tongji Hospital, Wuhan, China. IHH was diagnosed according to standard diagnostic criteria (1), and adult-onset IHH (AOIHH) was diagnosed based on previous published criteria (13). The study protocol (IRB ID: 20150302) (including collection of blood samples) was approved by the Ethics Committee of Huazhong University of Science and Technology in compliance with the Declaration of Helsinki (as revised in 2013). Informed consent was signed with each patient.
Panel sequencing and frequency analysis
Genomic DNA was extracted from peripheral blood lymphocytes in all patients. To focus on the genetic variations in IHH-associated genes, we designed a tailored next-generation panel which included 31 causal genes and 52 candidate genes, as described in our previous study (14). The detected gene loci included exon and proximal intron DNA sequences ≤15 bp from splice site. Non-synonymous rare sequencing variants (RSVs) with minor allele frequency <1% (ESP6500, 1000genome, gnomAD databases) were included in our study. Sanger sequencing was performed to confirm all variants by using an ABI PRISM 3130 genetic analyzer (Applied Biosystems, Foster City, CA, USA). The primers were as follows: GNRHR-forward: 5'-GACAGCTCTGGACAGACAAA-3'; GNRHR-reverse: 5'-ACACTAACTCTAAGGAATACATACCG-3'. Variants were reported in agreement with Human Genome Variation Society nomenclature (15).
Clinical evaluation
At first hospital visit, we performed detailed phenotyping and collected the medical history for each patient. Olfactory function was assessed by the olfactory testing or self-reporting. Tanner stage was evaluated by a senior physician according to Five Tanner stages of pubertal development (16). We defined testicular volume (TV) as the mean value of bilateral testicular volume measured by a Prader orchidometer. Serum hormone level [including follicle-stimulating hormone (FSH), luteinizing hormone (LH), total testosterone (T)] was determined before 9 a.m. Our patients were administered with human chorionic gonadotropin (hCG, Livzon Pharmaceutical Co., Zhuhai, China) due to financial reasons and/litter fertility desire. The dosage of hCG injection was 2,000 IU, twice per week. It would be adjusted according to serum T in the first 6 months. After 6 months, follow-up interval ranged from 3–6 months. Their Tanner stage, TV, serum T were recorded. Semen examination was recommended if patients were capable of masturbating. Semen examination complied to the rules of the World Health Organization laboratory manual for the examination and processing of human semen (fifth version) (17).
Conservative analysis and crystal structure prediction
The conservativeness of residue p.R240 in GNRHR protein was evaluated by sequence alignment in different vertebrates (Homo sapiens, Mus musculus, Rattus norvegicus, Bos taurus, Ovis aries, Cairina moschata and Danio rerio). By using a classic bioinformatic online tool, I-TASSER modeling (https://zhanglab.ccmb.med.umich.edu/I-TASSER/), we predicted crystal structure of GNRHR protein (both wild type and mutant type). Pattern diagram was supported by VMD software (the Beckman Institute, Illinios, USA).
Plasmids and GNRH decapeptide constructs
Plasmid (pEnter vector) of GNRHR-wild type (WT) expressing open reading frame of GNRHR (NM_000406.2) was obtained from Vigene Biosciences (Jinan, China). GNRHR-WT was fused with a C terminal Flag tag. We utilized the Quikchange II Site-Directed Mutagenesis kit (Stratagene, San Diego, CA, USA) to generate a plasmid of GNRHR-mutant type (MUT) (containing the GNRHR rare sequencing variant, c.719G>A, p.R240Q) based on the template plasmid. Given that function of decapeptide EHWSYGLRPG was identical to native GnRH sequence (18), we used the decapeptide obtained from Qiangyao Biosciences (Shanghai, China) to perform functional analysis.
Cell culture and transfection
Due to deficiency of endogenous GNRHR expression, Simian virus 40 (SV40)-infected CV1 (COS-7) cells (African green monkey kidney fibroblast-like cell line, ATCC, CRL-1651) was chosen as cell model in our study (19-21). COS-7 cells were seeded at standard 6 well dishes and cultured with complete media (10% fetal bovine serum). Two microgram vector was transfected into COS-7 cells using Lipofectamine® 3000 Reagent (Invitrogen, Shanghai, China) at 70–80% confluency. To exclude influence of transfection, we also transfected an empty plasmid as the MOCK group.
Real-time PCR
Total RNA from transfected COS-7 cells (the GNRHR-WT, GNRHR-MUT and MOCK groups) was extracted and used for cDNA synthesis by using Rever Tra Ace® qPCR RT kit (Toyobo, Osaka, Japan). We performed Real-time PCR based on an MX3000P quantitative PCR system (Agilent, Santa Clara, CA, USA) by using SYBR® Green Realtime PCR Master Mix (Toyobo, Osaka, Japan). We performed 40 cycles of denaturing at 95 °C for 15 s, followed by annealing at 60 °C for 15 s and extension at 72 °C for 45 s. GAPDH was used as an endogenous reference. Primer sequences were as follows: GNRHR-forward: 5'-GCACGGCTGAAGACTCTAAA-3'; GNRHR-reverse: 5'-AGGCAAAGAGAAAGAAGAAGTGA-3'; GAPDH-forward: 5'-CCATCTTCCAGGAACGAGAT-3'; GAPDH-reverse: 5'-GTTGCTGACGATCTTGAGGC-3'. 2−ΔΔCt analysis was conducted to calculate the mRNA level of GNRHR.
Western blots
After 24 h of transfection, protein samples were collected from the GNRHR-WT, GNRHR-MUT and MOCK groups. Cells were lysed and centrifuged at 12,000 rpm for 5 mins, and protein concentration was measured. Protein samples (25 microgram) were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (10%), and then transferred to nitrocellulose membranes. After blocking for 1 h, membranes were incubated with primary antibodies at 4 °C overnight. Herein, we used rabbit polyclonal Flag (Signalway Antibody, 20543-1-AP, Baltimore, MD, USA) to specifically detect GNRHR fused with Flag-tag (expressed from plasmid constructs). Other primary antibodies included rabbit monoclonal ERK1/2 (Cell Signaling Technology, 137F5, Danvers, MA, USA), rabbit monoclonal p-ERK1/2Thr202/Tyr204 (Cell Signaling Technology, 197G2), mouse monoclonal GAPDH (Proteintech, 60004-1-Ig, Wuhan, China). Membranes were washed and then incubated with secondary antibodies, goat anti-rabbit (Boster, BA1055, Wuhan, China) or goat anti-mouse (Boster, BA1051), for 1 h at room temperature. Band Density was analyzed by Image-Pro plus software (Media Cybernetics, Rockville, MD, USA) with the corresponding GAPDH as control.
ERK1/2 activation analysis
We first examined ERK1/2 activation after GnRH decapeptide (1 µM) stimulation (5 mins, 10 mins, 15 mins, 30 mins) in the GNRHR-WT group. The maximal response, defined as the highest level of p-ERK1/2/ERK1/2, was obtained at 5 mins, This time point was therefore chosen to perform further analysis of ERK1/2 activation. Transfected COS-7 cells were treated with GNRH decapeptide for 5 mins in the GNRHR-WT, GNRHR-MUT and MOCK groups. Protein samples were collected as previously described, and then expression of ERK1/2 and p-ERK1/2 was assessed by Western blots.
Inositol triphosphate determination
Transfected COS-7 cells were stimulated with GNRH decapeptide at concentration of 10-9 M, 10-8 M, 10-7 M, 10-6 M, 10-5 M for 1 h. Cell lysate was collected, and then supernatant was obtained after centrifugation at 12,000 rpm for 5 mins. We measured total inositol triphosphate (IP3) concentration by using the general IP3 ELISA kit (MyBioSource, San Diego, CA, USA) according to the standard protocol. Absorbance at 450 nm was examined with a microplate reader.
Intracellular calcium measurement
We treated transfected COS-7 cells with GnRH decapeptide (1 µM) for 1 h as previously described (21). Medium containing GnRH decapeptide was removed, and cells were washed three times with HBSS solution (HyClone, Shanghai, China). Cells were then incubated in 4 µM Fluo-4 AM solution (Yeasen, Shanghai, China) for 30 mins at 37 °C. Afterwards, cells were washed three times, and then incubated in HBSS solution for another 30 mins. Nuclear was stained with 6-diamidino-2-phenylindole (DAPI, Boster). Five microscopic images were randomly collected using a BX51 microscope (Olympus, Tokyo, Japan). Positive area of calcium was calculated using Image-Pro plus software.
Statistical analysis
Data analysis was performed using GraphPad Prism (GraphPad Software, San Diego, CA, USA). All the biological experiments were repeated three times independently. Chi-Square or Fisher’s Exact test was used to compare the allele frequencies of variants in different cohorts. Student's t-tests or one-way analysis of variance was used for normally distributed data, while nonparametric tests were used for nonnormally distributed data. normally distributed data were presented as mean ± standard deviation (SD), and nonnormally distributed data were presented as range or interquartile range. P values <0.05 were considered statistically significant.
Results
Mutation profile in IHH patients
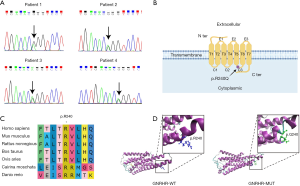
Of the cohort of 153 IHH patients, four sporadic patients were identified to have the same heterozygous p.R240Q rare sequencing variant (RSV, MAF <1%) in GNRHR gene (14) (Figure 1A). The prevalence of this RSV was 2.6% (4/153), while no other GNRHR RSVs were found in this cohort. Additionally, these four patients also harbored additional variants of IHH causal or candidate genes. We found PROKR2 and CNTN2 RSVs in Patient 1, SEMA7A RSV in Patient 2, TYRO3 RSV in Patient 3, as well as AXL and PLXNB1 RSVs in Patient 4. All RSVs were missense and heterozygous variants. Taking into account the patient's ethnicity (East Asian), we compared variant frequency between our patient cohort and the East Asian cohort by using the gnomAD database. The results indicated that allele frequencies of the GNRHR, PROKR2, and PLXNB1 RSVs were significantly higher in our patient cohort compared with East Asian cohort (P<0.001). However, no remarkable differences were found in frequencies of CNTN2, SEMA7A, TYRO3, and AXL RSVs (Table 1). These results suggest that the GNRHR RSV occurred more frequently in this Chinese IHH patient cohort than in the general population.
Full table
Clinical characteristics and outcomes
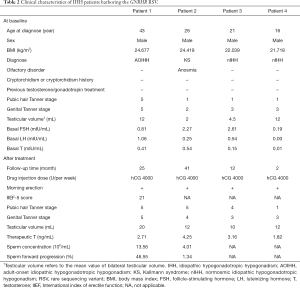
IHH is generally diagnosed during adolescence or early adulthood (1). As shown in Table 2, three of our four patients were diagnosed between the ages of 16 to 26 years. However, Patient 1 (43 years) was diagnosed with adult-onset IHH, since he exhibited symptoms of GnRH deficiency after sexual maturity. After detailed phenotyping, we found that Patient 2 showed anosmia but no additional phenotypes (including cryptorchidism, micropenis, cleft lip/palate, dental agenesis, optic nerve hypoplasia, sensorineural hearing loss, etc.). With exception of Patient 2, all patients in our cohort showed testicular volume (TV) >4 mL, which suggested partial puberty development. hCG treatment significantly improved their Tanner stage, TV, serum T levels. Additionally, all patients reported the resumption of morning erections after hCG treatment. Two patients exhibited seminal spermatozoa appearance after follow-up of more than two years. No side effects (acne and gynecomastia) were observed among these patients.
Full table
Bioinformatic and expression analysis of GNRHR RSV
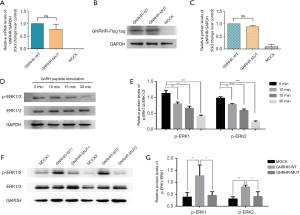
GNRHR is a member of the seven-transmembrane, G protein coupled receptor (GPCR) family in the cell membrane, and the GNRHR RSV is located in the third cytoplasmic loop of GNRHR (Figure 1B). Conservative analysis revealed that residue of p.R240 was highly conserved across seven different vertebrates (Figure 1C). In crystal structure prediction, replacement of arginine (strongly basic) with glutamine (neutral) changed the polarity of residue. The structure of this site has changed from alpha helix to curl. Glutamine has four fewer hydrogen atoms and two fewer nitrogen atoms (Figure 1D), which means that the replacement likely altered interactions with amino acids at other sites, and may further impair their function. Expression analysis showed that mRNA level of GNRHR in the GNRHR-WT group was mildly higher than that in GNRHR-MUT group (containing the GNRHR RSV), but the difference was not significant. Similarly, no significant difference was observed in protein comparison (Figure 2A,B,C).
GNRHR RSV affected ERK1/2 activation
One of the major signal transduction pathways of GNRHR is the MAPK cascade (9,22,23). To figure out the effect of GNRHR RSV on ERK1/2 activation, we first set different time points to find the optimal time of activation in the GNRHR-WT group. As shown in Figure 2D,E, the level of p-ERK1/2/ERK1/2 was highest at 5 min relative to 10 min, 15 min and 30 min. The level of p-ERK1/2/ERK1/2 gradually decreased as time elapsed. We subsequently used 5 mins for the duration of stimulation. Compared with the GNRHR-WT group, the level of p-ERK1/2/ERK1/2 was markedly lower in the GNRHR-MUT group (P<0.05, Figure 2F,G). The data thereby indicate that the GNRHR RSV impaires ERK1/2 activation to some extent.
GNRHR RSV affected IP3/calcium pathway
Because of IP3-mediated calcium mobilization in downstream of GNRHR (9,21), we examined IP3 accumulation and calcium mobilization to analyze the downstream effects of GNRHR RSV. As for IP3 accumulation, there was a remarkable difference between the GNRHR-WT and GNRHR-MUT groups (P<0.001), with the concentration of the GnRH decapeptide ranging from 10-8 M to 10-5 M (Figure 3A). For calcium mobilization, the highest calcium level was shown in the GNRHR-WT group. Calcium level partially, but significantly decreased in the GNRHR-MUT group than the GNRHR-WT group (P<0.05) (Figure 3B,C). Therefore, our result suggests that the GNRHR RSV partly damages IP3/calcium signal.

Discussion
In this study, we described a RSV in GNRHR gene and associated clinical manifestation. We identified four sporadic patients carrying the same heterozygous GNRHR RSV (p.R240Q). The prevalence of this GNRHR RSV was 2.6%. Its allele frequency was found to be exceptionally higher than that in general population. Functional analysis has demonstrated that this RSV caused loss of function (LOF) to GNRHR protein. It is of value to investigate the effect of the RSV, although the heterozygous state may be a non-functional factor in human, it homozygous/compound state could possibly have a pathogenic effect.
Heterozygous changes in GNRHR gene were enriched in IHH patients compared with controls. A large genetic screening for 863 probands with different forms of GnRH deficiency found that, 24 patients with monoallelic GNRHR mutations featured a broad phenotypic spectrum, ranging from Kallmann syndrome/nIHH, to attenuated GnRH deficiency (AOIHH, hypothalamic amenorrhea, or constitutional delay of puberty) (12). Intriguingly, researchers revealed that the heterozygous p.Q106R and R262Q RSVs were verified to be partial loss-of-function, and the heterozygous R139H RSV was complete loss-of-function (6,12). A recent study showed that two KS patients and one nIHH patient carried this heterozygous p.Q106R RSV, and they did not carry any other 36 IHH-associated gene variants (11). Therefore, these evidence indicates that heterozygous changes in GNRHR are not just a non-functional factor, but may lead to reproductive pathogenicity (11). Given the autosomal recessive inheritance, our study carefully suggests that the p.R240Q RSV found in four IHH patients may play a limited role in the process of morbidity. One possibility is that other mutations with unknown function on these patients caused the disease to occur. An additional possibility is that decreased gene dosage caused by monoallelic GNRHR mutation, makes individuals susceptible to environmental, behavioral, or psychosocial burdens on GnRH secretion (11,24).
Although GNRHR mutations were reported in normosmic IHH patients (25), we have demonstrated that the four patients showed phenotypic diversity. Patient 1, 3 and 4 had normal sense of smell, whereas Patient 2 were diagnosed with KS (containing olfactory disorder). Patient 1 displayed GnRH deficiency in adulthood, while the other three patients displayed that in pubertal development. At baseline, there were different levels of testicular development and hormone profiles. Testicular volume of Patient 3 and 4 were both above >4 mL, suggesting partial spontaneous puberty development (26), whereas TV of Patient 2 was 2 ml. Moreover, serum FSH levels of Patient 2 and 3 were at the lower level of normal range, but the rest two patients did not showed that. Consistently, other studies also proved this phenotypic diversity in patients with GNRHR mutations, ranging from complete IHH to constitutional delay of puberty (7,12,21).
Digenic and oligogenic inheritance are commonly observed in IHH (14,27). Herein, four patients with the GNRHR RSV harbored additional genetic abnormalities, including known IHH causal genes PROKR2, SEMA7A, AXL RSVs, and candidate genes CNTN2, TYRO3, and PLXNB1 RSVs. Except for the defined pathogenicity of PROKR2 RSV (p.W178S) (28,29), the role of remaining RSVs are currently not well understood. Of note, all four patients in our cohort showed good responses to hCG treatment. After follow-up of over 2 years, fertility outcomes of two patients were improved, with sperm concentration of 13.56 and 4.01×106/mL, respectively. Early studies indicated that TV >4 mL was a positive predictive factor for sperm production after receiving gonadotropin treatment (1,30,31). Given that the rest two patients also had TV >4 mL, it is plausible that hCG treatment may induce spermatogenesis in further follow-up.
It is well established that GnRH receptor is a member of the G protein coupled receptors (GPCRs), and highly expressed in the anterior pituitary gland, breast, ovary, and prostate (32). Mutations in GPCRs disrupt a variety of physiological processes, including protein biosynthesis, folding and transport, ligand binding, receptor activation, and interaction with G protein (10,33). Seventy-five percent of GNRHR variants reside in transmembrane domains (10). Our genetic analysis found that the RSV (p.R240Q) was located in the third cytoplasmic loop of GNRHR. As described in earlier studies (6,10,34-36), there are two more GNRHR variants (p.R262Q and p.L266R) reported in this loop. These variants both disrupted normal GNRHR expression and downstream signals. However, in our study, the p.R240Q RSV had no effect on expression, but negatively affected downstream signals, strongly suggesting that pathogenicity of this RSV may be milder than that of p.R262Q and p.L266R for the GNRHR protein.
According to severity of impaired function, we divided GNRHR variants into two subtypes. Complete loss-of-function (cLOF) is defined as no measurable in vitro signaling, while partial loss-of-function (pLOF) is defined when maximal signaling intensity is at least 10% of wild-type signaling (10). Protein analysis showed that the level of ERK1/2 phosphorylation in the GNRHR-MUT group was about 36–50% of that in the GNRHR-WT group. IP3 accumulation and calcium mobilization of GNRHR-MUT was approximately 20–54% of GNRHR-WT. Therefore, the GNRHR RSV is classified as a pLOF mutation. Moreover, our finding suggests that pLOF mutation may be associated with mild phenotypic forms, such as partial testicular development and normal FSH levels.
There are several limitations in this study. First, due to the geographic and economic reasons, it is difficult to obtain blood samples from patients’ family and ascertain their genotypes. Second, each patient harbored other gene mutations, we could not exclude the role of other possible pathogenic variants in the pathogenesis of IHH. Third, the intervals of follow-up review ranged from 3 to 6 months. A shorter and more consistent follow-up would be better for clinical management. Additionally, knowledge of IHH genes is rapidly progressing, our panel sequencing needs to be updated and more new causal genes should to be added into the panel.
Conclusions
In conclusion, the present study supports the loss of function of the GNRHR RSV (p.R240Q), and further expands its mutation and phenotypic spectrum. Monoallelic defects in GNRHR was found in Chinese IHH cohort, and it should be paid attention in this research field.
Acknowledgments
The authors thank every individual for their trust, cooperation, and contribution to this research. Yinwei Chen herein especially wants to thank Zhao Wang and Christina Wang for their great support.
Funding: This study was supported by grants from the National Natural Science Foundation of China (Project No. 81671443, 81601270).
Footnote
Reporting Checklist: The authors have completed the MDAR checklist. Available at http://dx.doi.org/10.21037/tau-20-1390
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tau-20-1390
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tau-20-1390). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study protocol (IRB ID: 20150302) (including collection of blood samples) was approved by the Ethics Committee of Huazhong University of Science and Technology in compliance with the Declaration of Helsinki (as revised in 2013). Informed consent was signed with each patient.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Boehm U, Bouloux PM, Dattani MT, et al. Expert consensus document: European Consensus Statement on congenital hypogonadotropic hypogonadism--pathogenesis, diagnosis and treatment. Nat Rev Endocrinol 2015;11:547-64. [Crossref] [PubMed]
- Bhagavath B, Podolsky RH, Ozata M, et al. Clinical and molecular characterization of a large sample of patients with hypogonadotropic hypogonadism. Fertil Steril 2006;85:706-13. [Crossref] [PubMed]
- Mitchell AL, Dwyer A, Pitteloud N, et al. Genetic basis and variable phenotypic expression of Kallmann syndrome: towards a unifying theory. Trends Endocrinol Metab 2011;22:249-58. [Crossref] [PubMed]
- Maione L, Dwyer AA, Francou B, et al. GENETICS IN ENDOCRINOLOGY: Genetic counseling for congenital hypogonadotropic hypogonadism and Kallmann syndrome: new challenges in the era of oligogenism and next-generation sequencing. Eur J Endocrinol 2018;178:R55-80. [Crossref] [PubMed]
- Young J, Xu C, Papadakis GE, Acierno JS, et al. Clinical Management of Congenital Hypogonadotropic Hypogonadism. Endocr Rev 2019;40:669-710. [Crossref] [PubMed]
- de Roux N, Young J, Misrahi M, et al. A family with hypogonadotropic hypogonadism and mutations in the gonadotropin-releasing hormone receptor. N Engl J Med 1997;337:1597-602. [Crossref] [PubMed]
- Bhagavath B, Ozata M, Ozdemir I, et al. The prevalence of gonadotropin-releasing hormone receptor mutations in a large cohort of patients with hypogonadotropic hypogonadism. Fertil Steril 2005;84:951-7. [Crossref] [PubMed]
- Fan NC, Peng C, Krisinger J, et al. The human gonadotropin-releasing hormone receptor gene: complete structure including multiple promoters, transcription initiation sites, and polyadenylation signals. Mol Cell Endocrinol 1995;107:R1-8. [Crossref] [PubMed]
- Sperduti S, Limoncella S, Lazzaretti C, et al. GnRH Antagonists Produce Differential Modulation of the Signaling Pathways Mediated by GnRH Receptors. Int J Mol Sci 2019;20:5548. [Crossref] [PubMed]
- Cioppi F, Riera-Escamilla A, Manilall A, et al. Genetics of ncHH: from a peculiar inheritance of a novel GNRHR mutation to a comprehensive review of the literature. Andrology 2019;7:88-101. [Crossref] [PubMed]
- Amato LGL, Montenegro LR, Lerario AM, et al. New genetic findings in a large cohort of congenital hypogonadotropic hypogonadism. Eur J Endocrinol 2019;181:103-19. [Crossref] [PubMed]
- Gianetti E, Hall JE, Au MG, et al. When genetic load does not correlate with phenotypic spectrum: lessons from the GnRH receptor (GNRHR). J Clin Endocrinol Metab 2012;97:E1798-807. [Crossref] [PubMed]
- Nachtigall LB, Boepple PA, Pralong FP, et al. Adult-onset idiopathic hypogonadotropic hypogonadism--a treatable form of male infertility. N Engl J Med 1997;336:410-5. [Crossref] [PubMed]
- Zhou C, Niu Y, Xu H, et al. Mutation profiles and clinical characteristics of Chinese males with isolated hypogonadotropic hypogonadism. Fertil Steril 2018;110:486-95.e5. [Crossref] [PubMed]
- den Dunnen JT, Antonarakis SE. Mutation nomenclature extensions and suggestions to describe complex mutations: a discussion. Hum Mutat 2000;15:7-12. [Crossref] [PubMed]
- Marshall WA, Tanner JM. Variations in the pattern of pubertal changes in boys. Arch Dis Child 1970;45:13-23. [Crossref] [PubMed]
- World Health Organization. WHO laboratory manual for the examination and processing of human semen. 5th ed. Geneva: World Health Organization, 2010.
- Zeng W, Pagnon J, Jackson DC. The C-terminal pentapeptide of LHRH is a dominant B cell epitope with antigenic and biological function. Mol Immunol 2007;44:3724-31. [Crossref] [PubMed]
- Maya-Núñez G, Janovick JA, Aguilar-Rojas A, et al. Biochemical mechanism of pathogenesis of human gonadotropin-releasing hormone receptor mutants Thr104Ile and Tyr108Cys associated with familial hypogonadotropic hypogonadism. Mol Cell Endocrinol 2011;337:16-23. [Crossref] [PubMed]
- Janovick JA, Ulloa-Aguirre A, Conn PM. Evolved regulation of gonadotropin-releasing hormone receptor cell surface expression. Endocrine 2003;22:317-27. [Crossref] [PubMed]
- Correa-Silva SR, Fausto JDS, Kizys MML, et al. A novel GNRHR gene mutation causing congenital hypogonadotrophic hypogonadism in a Brazilian kindred. J NeuroendocrinoL 2018;30:e12658 [Crossref] [PubMed]
- Naor Z, Benard O, Seger R. Activation of MAPK Cascades by G-protein-coupled Receptors: The Case of Gonadotropin-releasing Hormone Receptor. Trends Endocrinol Metab 2000;11:91-9. [Crossref] [PubMed]
- Kawaminami M, Uematsu N, Funahashi K, et al. Gonadotropin releasing hormone (GnRH) enhances annexin A5 mRNA expression through mitogen activated protein kinase (MAPK) in LbetaT2 pituitary gonadotrope cells. Endocr J 2008;55:1005-14. [Crossref] [PubMed]
- Marcus MD, Loucks TL, Berga SL. Psychological correlates of functional hypothalamic amenorrhea. Fertil Steril 2001;76:310-6. [Crossref] [PubMed]
- Beranova M, Oliveira LM, Bedecarrats GY, et al. Prevalence, phenotypic spectrum, and modes of inheritance of gonadotropin-releasing hormone receptor mutations in idiopathic hypogonadotropic hypogonadism. J Clin Endocrinol Metab 2001;86:1580-8. [Crossref] [PubMed]
- Pitteloud N, Acierno JJ, Meysing A, et al. Mutations in fibroblast growth factor receptor 1 cause both Kallmann syndrome and normosmic idiopathic hypogonadotropic hypogonadism. Proc Natl Acad Sci U S A 2006;103:6281-6. [Crossref] [PubMed]
- Men M, Wu J, Zhao Y, et al. Genotypic and phenotypic spectra of FGFR1, FGF8, and FGF17 mutations in a Chinese cohort with idiopathic hypogonadotropic hypogonadism. Fertil Steril 2020;113:158-66. [Crossref] [PubMed]
- Cole LW, Sidis Y, Zhang C, et al. Mutations inProkineticin 2 and Prokineticin receptor 2 genes in Human Gonadotrophin-Releasing Hormone Deficiency: Molecular Genetics and Clinical Spectrum. J Clin Endocrinol Metab 2008;93:3551-9. [Crossref] [PubMed]
- Monnier C, Dodé C, Fabre L, et al. PROKR2 missense mutations associated with Kallmann syndrome impair receptor signalling activity. Hum Mol Genet 2009;18:75-81. [Crossref] [PubMed]
- Chen Y, Sun T, Niu Y, et al. Correlations Among Genotype and Outcome in Chinese Male Patients With Congenital Hypogonadotropic Hypogonadism Under HCG Treatment. The Journal of Sexual Medicine 2020;17:645-57. [Crossref] [PubMed]
- Liu PY, Baker HWG, Jayadev V, et al. Induction of Spermatogenesis and Fertility during Gonadotropin Treatment of Gonadotropin-Deficient Infertile Men: Predictors of Fertility Outcome. The Journal of Clinical Endocrinology & Metabolism 2009;94:801-8. [Crossref] [PubMed]
- Aguilar-Rojas A, Huerta-Reyes M, Maya-Nunez G, et al. Gonadotropin-releasing hormone receptor activates GTPase RhoA and inhibits cell invasion in the breast cancer cell line MDA-MB-231. BMC Cancer 2012;12:550. [Crossref] [PubMed]
- Tao YX, Conn PM. Chaperoning G protein-coupled receptors: from cell biology to therapeutics. Endocr Rev 2014;35:602-47. [Crossref] [PubMed]
- Choi JH, Balasubramanian R, Lee PH, et al. Expanding the Spectrum of Founder Mutations Causing Isolated Gonadotropin-Releasing Hormone Deficiency. J Clin Endocrinol Metab 2015;100:E1378-85. [Crossref] [PubMed]
- Maya-Núñez G, Janovick JA, Ulloa-Aguirre A, et al. Molecular basis of hypogonadotropic hypogonadism: restoration of mutant (E(90)K) GnRH receptor function by a deletion at a distant site. J Clin Endocrinol Metab 2002;87:2144-9. [Crossref] [PubMed]
- Flanagan CA, Rodic V, Konvicka K, et al. Multiple interactions of the Asp(2.61(98)) side chain of the gonadotropin-releasing hormone receptor contribute differentially to ligand interaction. Biochemistry 2000;39:8133-41. [Crossref] [PubMed]


