
Genomic analysis of response to bacillus Calmette-Guérin (BCG) treatment in high-grade stage 1 bladder cancer patients
Introduction
The annual incidence of urothelial carcinoma of the bladder (UCB) in the United States is estimated to reach more than 83,000 cases in 2020, making it the 6th most common non-cutaneous malignancy (1). Non-muscle invasive bladder cancer (NMIBC) consists of carcinoma in situ (CIS), Ta, and T1 tumors and accounts for approximately 75% of new diagnoses (2). Overall, NMIBC carries excellent long-term prognosis with a treatment approach focusing on local resection with or without intravesical therapy (3). Tumors with the highest risk for progression (15% at 5 years) to muscle invasive bladder cancer (MIBC) are the subset including high-grade (HG) T1 disease, having demonstrated invasive biology by disrupting the lamina propria. Standard front-line therapy for high-risk NMIBC is visually complete transurethral resection (TUR) plus intravesical bacillus Calmette-Guérin (BCG) administration, but recurrence is common at 40% within 1 year of treatment and there remains a 5-year cancer specific mortality rate of 25% (4,5).
The mechanism of action for BCG therapy is believed to involve immune activation and destruction of local cancer cells but the phenomenon is poorly understood (6). There are no predictors for treatment response outside of classical clinicopathologic features, and the high treatment failure rate highlights the need for additional measures. In the current era of next generation sequencing (NGS) for UCB, much research effort to date has been focused on exploring the prognostic and predictive role for MIBC genomics as it pertains to treatment response, with relatively less attention paid to NMIBC genomics and BCG responsiveness on a post hoc basis (7-10).
There remains ample opportunity and need for NMIBC biomarkers associated with BCG response to improve patient selection for intravesical therapy versus upfront surgery. Furthermore, comparisons between responders and non-durable responders might lead to a better mechanistic understanding of BCG therapy. We used NGS (RNAseq and targeted DNA sequencing) to analyze a cohort of BCG treated NMIBC patients with the highest risk of disease progression (HG T1) stratified by treatment response to compare differences in expressional and mutational profiles in relation to BCG responsiveness. We present the following article in accordance with the MDAR reporting checklist (available at https://dx.doi.org/10.21037/tau-21-158).
Methods
Patient characteristics
Recognizing the difficulties with clinical heterogeneity in NGS studies, we consciously elected to a priori restrict our analysis to patients with HG T1 UCB treated with complete induction BCG (≥5 of 6 weekly instillations) and compare those who responded to therapy versus those who did not. We retrospectively identified such patients at least 18 years or older at our institution between 2007 and 2019. Patients were excluded from further analysis if they had received any BCG within the previous 1 year. Response to BCG therapy was then assessed for each patient in order to create two discrete categories: BCG non-durable responders and BCG durable responders. Using date of first induction BCG instillation as the reference point, BCG non-durable responders were defined as patients with recurrence of UCB (any stage or grade) within 2 years, and BCG durable responders had no recurrences detected at any point during observable follow-up with a minimum disease-free interval of at least 2 years. BCG non-durable responders included those with a partial response, defined as any recurrence of lower grade or stage than the index lesion. Maintenance BCG was administered in all eligible patients in accordance with accepted guidelines (11). Formalin-fixed paraffin-embedded (FFPE) tissue blocks from the diagnostic TUR immediately preceding induction BCG were available for 45 patients. After microdissection and purification of DNA and RNA, a total of 42 samples were available for analysis.
DNA sequencing and variant calling
DNA samples were sequenced using the Trusight Tumor 170 gene panel (TST170, Illumina Inc., San Diego, CA, USA). Three hundred and fifty paired-end reads in fastq file format were first checked for quality using FastQC, a freely available software. The reads were then trimmed of adapter sequences and low-quality bases using Trimmomatic (12) and checked for quality again following trimming. The reads were aligned to the human genome 38 (hg38) reference genome using the BWA MEM aligner (13). The alignment files were then condensed by lane to a single alignment per sample. These alignments were assessed for quality and depth of coverage using Picard HSMetrics tool (http://broadinstitute.github.io/picard/). The average depth of coverage was 580X, with a median depth of 375X. After being assessed for quality, the alignments were passed to the Genome Analysis Toolkit (GATK) for variant calling (14). A panel of normal references was created using genomes listed in the 1000 Genomes Project. The variants were also quality checked, filtered, and annotated using the GATK package. After variants were called, they were compared between groups (durable responders vs. non-durable responders) using a contingency table and a fisher’s exact test. All variant P values listed are raw P values and any meeting a cutoff P value <0.05 were selected for downstream analysis.
RNA sequencing, gene expression, and variant calling
RNA samples were sequenced using Illumina’s TruSeq RNA whole exome panel. Sequenced reads were quality checked and reads were aligned to the human genome hg38 reference set using STAR aligner (15). The aligned files were used for further analysis of gene expression and variant calling. Counts of mapped reads for the genes were calculated by featureCounts in the R package SubRead (16). These read counts were used as input for DESeq2 to identify significant differentially expressed genes between durable responders and BCG non-durable responder samples. After adjusting for multiple correction using the Benjamini-Hochberg method, those with an adjusted P value of less than 0.05 and log-fold change difference greater than two were considered statistically significant. These significantly differentially expressed genes were inputted into the Ingenuity Pathway Analysis (IPA) software (QIAGEN Inc., https://www.qiagenbioinformatics.com/products/ingenuity-pathway-analysis) to identify pathways that are potentially affected (17). Clinically annotated DNA sequencing and RNAseq data was uploaded to the National Center for Biotechnology Information Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/).
Statistical analysis
Read counts were normalized and log transformed in R for input into classifyT1BC (Meeks classifier), consensusMIBC, and NMIBC classification (UROMOL classifier) to identify classification subtypes within our dataset (10,18,19). The classifyT1BC and consensusMIBC classifier tools were run using their respective R packages, while the NMIBC classification was run by uploading the log transformed normalized counts at http://nmibc-class.dk. For RNA-seq variant calling, aligned reads were merged per sample and duplicates marked with Picard tools. Sequence recalibration from GATK tools were used as input to Mutect2 for variant calling. Since matched normal samples were not available, a panel of normal references was created with MuTect2 to filter out potential germline variants. Raw variant calls for samples were then used for downstream analyses. After all variants were called, they were passed to bcftools to pull common variants from all samples. Only variants present in at least three samples were included. These common variants were then assessed for significance between groups (durable responders vs. non-durable responders) using fisher’s exact test in R. Variants were filtered to only include those with a raw P value of <0.05 (none had an adjusted P value of <0.05). Further annotation was done on the common variants using the Annotate Variation (Annovar) software to predict amino acid changes (20). The significant common variants were further filtered to only nonsynonymous variants. These filtered variants were further filtered to those that were annotated to genes that were found to be differentially expressed. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by institutional/regional/national ethics/committee/ethics board of the Levine Cancer Institute of Atrium Health (No. LCI-GU-SPEC-PHA-001) and individual consent for this retrospective analysis was waived.
Results
Patient characteristics
A total of 42 patients were included in the final analysis, of which 17 (40.5%) were BCG durable responders and 25 (59.5%) were BCG non-durable responders. With a median overall follow-up of 51.7 months [interquartile range (IQR), 29.3–69.7 months] baseline characteristics of the cohort (Table 1) included a median age of 68.9 years old (IQR, 64.7–73.5 years old) comprised of 78.6% men (n=33), 78.6% current or former smokers (n=33), and 12.1% non-white ethnicities (n=5). A markedly higher proportion of females were observed among durable responders (n=7, 41.2%) than non-durable responders (n=2, 8.0%, P=0.019). Additionally, there were no active smokers among the durable responders, as compared to 24.0% (n=6, P=0.057) in the non-durable responder group.
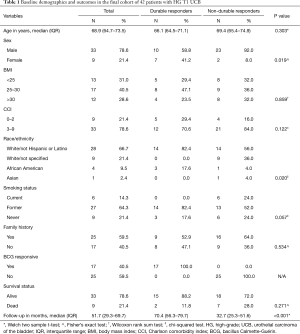
Full table
Recurrences in the non-durable responders (Table 2) occurred at a median of 7.7 months (IQR, 5.4–8.8 months) from initiation of induction BCG and were predominantly HG (n=22, 88.0%). Partial response was observed in 48.0% (n=12). There were 3 patients (12.0%) who progressed to muscle invasion or higher during follow-up, including a single patient who developed distant metastasis in the setting of negative surveillance cystoscopy.
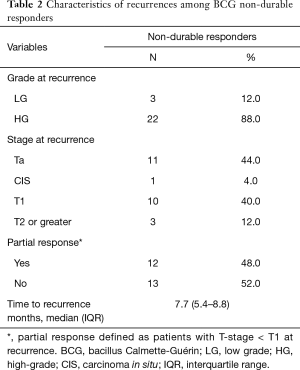
Full table
Gene expression
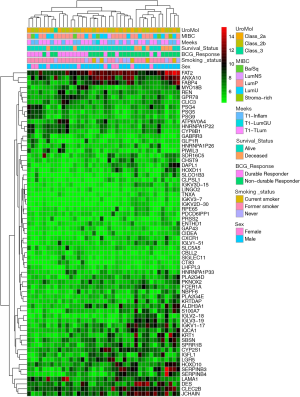
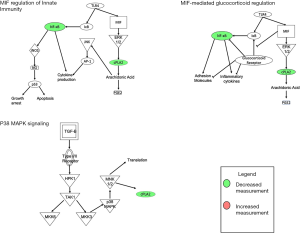
In total, 67 genes (Table S1) were found to have statistically significant (P<0.05) differential RNA expression on RNAseq when comparing durable responders to non-durable responders. The top genes found to be mutated across all samples, including both durable responders and non-durable responders, were BRCA2, ATM, and EP300 at 96.73%, 93.55%, and 90.32% respectively. The results of unsupervised hierarchical clustering analysis are shown in Figure 1 along with select clinicopathologic factors. Selected publicly available classification schemes for urothelial carcinoma were applied. To test whether these were involved in mediating inflammatory pathways, the differentially expressed genes were analyzed using the IPA software, using the expression levels for non-durable responders as the reference value. Three of the top pathways with statistically significant differences in gene expression (Table 3; Figure 2) were directly or indirectly associated with inflammation including macrophage migration inhibitory factor (MIF)-mediated glucocorticoid regulation (Figure S1), MIF regulation of innate immunity (Figure S2), and p38 mitogen activated protein family of kinases (MAPK) signaling (Figure S3). Furthermore, the predicted impact of changes in specific gene expression among durable responders was significant global up-regulation as well as activation of the key inflammatory mediator NF-κB when compared to non-durable responders (Table 3).

Full table
DNA and RNA variant analysis
In addition to gene expression, we analyzed the RNAseq dataset for variants that may be potentially useful biomarker candidates. The pool of RNA variants (Figure S4) was first filtered to variants that appeared in at least three samples (n=126,033 variants). These were further filtered to variants that were annotated to proteins as nonsynonymous mutations (n=44,798 variants), of which 856 variants were significantly enriched (raw P value <0.05). Finally, of these significant nonsynonymous variants, there were six variant genes identified (Figure 3A) from among the 228 genes previously identified as being differentially expressed based on BCG responsiveness. Notably, using these criteria, none of the differentially expressed genes from the pathway analysis were also RNA variants considered to be biomarker candidates. We found that JCHAIN variants were observed only in the non-durable responder samples (47.8% vs. 0%, P<0.001), while S100A7 (39.1% vs. 5.9%, P=0.026), CLEC2B (43.5% vs. 11.8%, P=0.041), and S100A7 (52.2% vs. 17.6%, P=0.046) were also significantly enriched in those patients (Figure 3B; Table S2).
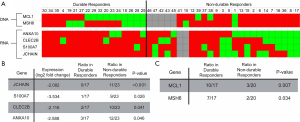
Analysis of DNA variants was conducted with the TST170 gene panel using similar iterative exclusion methodology as above (Figure S4), with 150 DNA variants initially identified in three or more tumor samples, of which 45 were annotated to proteins as nonsynonymous mutations. There was statistically significant (P value <0.05) enrichment of two DNA variants that also passed nonsynonymous filtration (Figure 3A). Each of the identified DNA variants were observed in a higher proportion of durable responders (Figure 3C; Table S3): frameshift deletions in MCL1 (58.8% vs. 15%, P=0.007) and MSH6 (41.2% vs. 10.0%, P=0.034).
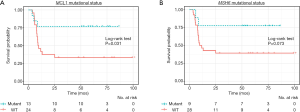
All DNA and RNA variants showing a statistically significant association with BCG response were then analyzed for impact in recurrence free survival (RFS) using the Kaplan-Meier survival method. A deleterious mutation in the MCL1 gene (Figure 4A) was associated with significantly prolonged RFS as compared to wild type (2-year RFS 77.0% vs. 37.5%; log-rank test P=0.031). Though not reaching predefined statistical significance, mutation of the MSH6 gene (Figure 4B) was also associated with longer 2-year RFS (77.8% vs. 42.9%; log-rank test P=0.073).
Discussion
Predicting BCG responsiveness in NMIBC has remained an elusive goal, even as NGS techniques have vastly increased availability of genomic data. We sought to identify potential biomarkers for predicting response using a cohort of patients which were a priori selected to have similar baseline pathologic features (HG T1 tumors) who were treated with “adequate” BCG and displayed either durable complete response (median follow-up 70.4 months; IQR, 56.3–79.7) or disease relapse before 2 years of follow-up (median time to recurrence 7.7 months; IQR, 5.4–8.8). In this cohort, deleterious mutations in the MCL1 gene were associated with significantly longer RFS (median 9.3 months vs. not reached; P=0.031). Additional pathway analysis identified notable inflammatory pathways linked to durable responders with predicted increased pathway activation and activation of NF-κB.
MCL1 is an anti-apoptotic protein of the BCL2 family and its down-regulation has been linked to tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) sensitization (6,21). In prior work using immunohistochemical staining for BCL2, expression was shown to be inversely correlated with recurrence of NMIBC following BCG treatment with 47% prevalence at baseline in patients with tumor recurrences following BCG therapy as compared to 79% staining positive among non-recurrent cases (22). TRAIL expression has been shown to be up-regulated in granulocytes following induction of BCG and release of interferon signaling particles by cancer cells, and is therefore believed to be a key death ligand in the anti-neoplastic properties of BCG (23). The location of the frameshift deletion within MCL1 we detected falls in the first exon near the N-terminus within a region believed to be involved in protein regulation (24). We speculate, therefore, that sensitization to TRAIL, induced by a loss of function mutation in MCL1 that is seen more frequently in the durable responders and in association with longer RFS, provides a mechanism by which BCG responsiveness could be enhanced. The combination of a plausible mechanistic explanation for the association between MCL1 and BCG responsiveness makes this a promising target for further biomarker validation.
Deleterious mutations in the DNA mismatch repair protein MSH6 were also closely linked to longer RFS, though this did not meet our prespecified cutoff for statistical significance (P=0.073), a fact that may be explained by the small sample sizes. The MSH6 gene is well known for its role in Lynch syndrome whereby germline mutations lead to accumulation of errors in regions of DNA called microsatellites. The classic genitourinary manifestation of Lynch syndrome is that of upper tract urothelial carcinoma which is related to UCB through a shared histologic origin, but which has significant differences in genomic signatures (25). Beyond the association with Lynch syndrome, MSH6 has been linked specifically to UCB where methylation status of MSH6 has shown an association with progression and recurrence among patients treated with intravesical BCG (26). The deactivating, hypermethylated state of MSH6 corresponded to an increased time to progression and improved survival outcomes following BCG therapy in patients with HG NMIBC, and as such, should be considered for further investigation into its potential for a biomarker of response to therapy.
The pathways identified in our gene expression analysis show that inflammation and immune response are important triggers for BCG response, which we confirm in our formal pathway analysis using IPA software. MIF-mediated glucocorticoid regulation and MIF regulation of innate immunity, two of the top pathways from our analysis, are strongly involved in the immune system response. MIF is released during inflammatory reactions and has been shown to promote the expression of several pro-inflammatory cytokines (27). Upregulation of MIF-related pathways predicted by IPA software (Figures S1,S2) that we observed in our samples would be expected to result in a pro-inflammatory environment conducive to immune-mediated tumor destruction. Interestingly, there is a common linkage between all three of these pathways in the form of decreased expression in the NFKB1 gene leading to upregulation of each associated pathway (Table 3). Functionally, the NFKB1 gene is involved in regulating transcription for many pathways associated with inflammation, immunity, and apoptosis (28).
Other variants identified in our cohort, CLEC2B and S100A7, can also be linked to NF-κB activity but the correlation is less direct. CLEC2B is a member of the C-type lectin superfamily of proteins, carbohydrate binding proteins that can also self-bind and modulate many physiological activities (29). Increased TRAIL levels have been linked to C-type lectin-like molecule binding, but no specific work has shown direct correlation with expression of the CLEC2B gene. S100A7 is a less well described pro-survival protein present in other cancer models (30). It has also been linked to NF-κB signaling through suppression of the Yes-associated protein (YAP) oncogene cascade (31) and can be evaluated with immunohistochemical staining, though a previous study failed to find statistically significant association with BCG response (32). Though speculative until confirmed by further study, these observations lend additional support to support the hypothesis that alterations in NF-κB and its associated inflammatory regulatory networks may impact the response to BCG in patients with HG T1 UCB. ANXA10 is from the annexin family of proteins that are calcium dependent membrane binding proteins (33). While ANXA10’s specific function is still unknown, high expression levels have previously been linked to lower rates of NMIBC progression and improved survival outcomes (33,34).
The present study is limited by the small sample size available for genomic analysis, though we believe a strength of our data lies in the homogeneity of primary tumor samples. By narrowing our criteria to only HG T1 tumors that had clearly either responded to BCG, with a median disease-free follow-up of nearly 6 years, versus patients with relapse at a median of less than 8 months, we have intentionally maximized the difference in clinical outcomes. The use of a targeted 170 gene panel (Illumina’s TST170) is another potential limitation. While there may be additional interesting variants to identify through whole-exome sequencing, the targeted panel increases confidence as it provides higher sequencing coverage depth of the selected genes. Due to the retrospective nature of our dataset, we were not able to collect normal samples for matched analysis. Matching tumor/normal sample pairs are important in filtering out potential germline variants from the true somatic variants and represents an important future step in this work.
Conclusions
Intravesical BCG remains the preferred frontline treatment for most patients with HG T1 bladder cancer who have the highest risk of progression to muscle invasive disease. In this vulnerable patient population there is an ongoing need to identify reliable biomarkers that can predict patient responsiveness to BCG immunotherapy. We have identified several differentially mutated and expressed genes that vary based on responsiveness to therapy that can serve as candidate biomarkers to be further interrogated with validation studies, specifically MCL1 and MSH6. Furthermore, based on our work we hypothesize that NF-κB signaling may be a common thread linking these variants and pathways we analyzed. This serves as the foundation for further investigation into these alterations as possible rational biomarkers for high-risk UCB treatment.
Acknowledgments
The authors would like to acknowledge Illumina, Inc. for providing TruSight Tumor 170 kits and associated consumables for the targeted DNA sequencing portion of this study.
Funding: Carolinas Bladder Cancer Fund; Don and Betty Anderson Fund for Bladder Cancer Research.
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://dx.doi.org/10.21037/tau-21-158
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/tau-21-158). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by institutional/regional/national ethics/committee/ethics board of the Levine Cancer Institute of Atrium Health (No. LCI-GU-SPEC-PHA-001) and individual consent for this retrospective analysis was waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin 2020;70:7-30. [Crossref] [PubMed]
- Burger M, Catto JW, Dalbagni G, et al. Epidemiology and risk factors of urothelial bladder cancer. Eur Urol 2013;63:234-41. [Crossref] [PubMed]
- Cambier S, Sylvester RJ, Collette L, et al. EORTC nomograms and risk groups for predicting recurrence, progression, and disease-specific and overall survival in non-muscle-invasive stage Ta-T1 urothelial bladder cancer patients treated with 1-3 years of maintenance bacillus Calmette-Guérin. Eur Urol 2016;69:60-9. [Crossref] [PubMed]
- Chang SS, Bochner BH, Chou R, et al. Treatment of nonmetastatic muscle-invasive bladder cancer: American Urological Association/American Society of Clinical Oncology/American Society for Radiation Oncology/Society of Urologic Oncology Clinical Practice Guideline Summary. J Oncol Pract 2017;13:621-5. [Crossref] [PubMed]
- Dalbagni G, Vora K, Kaag M, et al. Clinical outcome in a contemporary series of restaged patients with clinical T1 bladder cancer. Eur Urol 2009;56:903-10. [Crossref] [PubMed]
- Redelman-Sidi G, Glickman MS, Bochner BH. The mechanism of action of BCG therapy for bladder cancer--a current perspective. Nat Rev Urol 2014;11:153-62. [Crossref] [PubMed]
- Hedegaard J, Lamy P, Nordentoft I, et al. Comprehensive transcriptional analysis of early-stage urothelial carcinoma. Cancer Cell 2016;30:27-42. [Crossref] [PubMed]
- Pietzak EJ, Bagrodia A, Cha EK, et al. Next-generation sequencing of nonmuscle invasive bladder cancer reveals potential biomarkers and rational therapeutic targets. Eur Urol 2017;72:952-9. [Crossref] [PubMed]
- van Kessel KEM, van der Keur KA, Dyrskjøt L, et al. Molecular markers increase precision of the European Association of Urology Non-Muscle-Invasive Bladder Cancer Progression Risk Groups. Clin Cancer Res 2018;24:1586-93. [Crossref] [PubMed]
- Robertson AG, Groeneveld CS, Jordan B, et al. Identification of differential tumor subtypes of T1 bladder cancer. Eur Urol 2020;78:533-7. [Crossref] [PubMed]
- Chang SS, Boorjian SA, Chou R, et al. Diagnosis and treatment of non-muscle invasive bladder cancer: AUA/SUO guideline. J Urol 2016;196:1021-9. [Crossref] [PubMed]
- Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 2014;30:2114-20. [Crossref] [PubMed]
- Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv, 2013:1303.3997.
- Van der Auwera GA, Carneiro MO, Hartl C, et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics 2013;43:11.10.1-11.10.33.
- Dobin A, Davis CA, Schlesinger F, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29:15-21. [Crossref] [PubMed]
- Liao Y, Smyth GK, Shi W. The R package Rsubread is easier, faster, cheaper and better for alignment and quantification of RNA sequencing reads. Nucleic Acids Res 2019;47:e47 [Crossref] [PubMed]
- Krämer A, Green J, Pollard J Jr, et al. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 2014;30:523-30. [Crossref] [PubMed]
- Kamoun A, de Reyniès A, Allory Y, et al. A consensus molecular classification of muscle-invasive bladder cancer. Eur Urol 2020;77:420-33. [Crossref] [PubMed]
- Lindskrog SV, Prip F, Lamy P, et al. An integrated multi-omics analysis identifies prognostic molecular subtypes of non-muscle-invasive bladder cancer. Nat Commun 2021;12:2301. [Crossref] [PubMed]
- Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 2010;38:e164 [Crossref] [PubMed]
- Kim SH, Ricci MS, El-Deiry WS. Mcl-1: a gateway to TRAIL sensitization. Cancer Res 2008;68:2062-4. [Crossref] [PubMed]
- Okamura T, Akita H, Kawai N, et al. Immunohistochemical evaluation of p53, proliferating cell nuclear antigen (PCNA) and bcl-2 expression during bacillus Calmette-Guerin (BCG) intravesical instillation therapy for superficial bladder cancers. Urol Res 1998;26:161-4. [Crossref] [PubMed]
- Ludwig AT, Moore JM, Luo Y, et al. Tumor necrosis factor-related apoptosis-inducing ligand: a novel mechanism for Bacillus Calmette-Guérin-induced antitumor activity. Cancer Res 2004;64:3386-90. [Crossref] [PubMed]
- Germain M, Duronio V. The N terminus of the anti-apoptotic BCL-2 homologue MCL-1 regulates its localization and function. J Biol Chem 2007;282:32233-42. [Crossref] [PubMed]
- Moss TJ, Qi Y, Xi L, et al. Comprehensive genomic characterization of upper tract urothelial carcinoma. Eur Urol 2017;72:641-9. [Crossref] [PubMed]
- Agundez M, Grau L, Palou J, et al. Evaluation of the methylation status of tumour suppressor genes for predicting bacillus Calmette-Guérin response in patients with T1G3 high-risk bladder tumours. Eur Urol 2011;60:131-40. [Crossref] [PubMed]
- Nobre CC, de Araújo JM, Fernandes TA, et al. Macrophage migration inhibitory factor (MIF): biological activities and relation with cancer. Pathol Oncol Res 2017;23:235-44. [Crossref] [PubMed]
- Li P, Gu J, Yang X, et al. Functional promoter -94 ins/del ATTG polymorphism in NFKB1 gene is associated with bladder cancer risk in a Chinese population. PLoS One 2013;8:e71604 [Crossref] [PubMed]
- Wiersma VR, de Bruyn M, Shi C, et al. C-type lectin-like molecule-1 (CLL1)-targeted TRAIL augments the tumoricidal activity of granulocytes and potentiates therapeutic antibody-dependent cell-mediated cytotoxicity. MAbs 2015;7:321-30. [Crossref] [PubMed]
- Emberley ED, Niu Y, Curtis L, et al. The S100A7-c-Jun activation domain binding protein 1 pathway enhances prosurvival pathways in breast cancer. Cancer Res 2005;65:5696-702. [Crossref] [PubMed]
- Li Y, Kong F, Shao Q, et al. YAP expression and activity are suppressed by S100A7 via p65/NFκB-mediated repression of ΔNp63. Mol Cancer Res 2017;15:1752-63. [Crossref] [PubMed]
- Kitamura H, Torigoe T, Honma I, et al. Effect of human leukocyte antigen class I expression of tumor cells on outcome of intravesical instillation of bacillus calmette-guerin immunotherapy for bladder cancer. Clin Cancer Res 2006;12:4641-4. [Crossref] [PubMed]
- Munksgaard PP, Mansilla F, Brems Eskildsen AS, et al. Low ANXA10 expression is associated with disease aggressiveness in bladder cancer. Br J Cancer 2011;105:1379-87. [Crossref] [PubMed]
- Jin S, Chang IH, Kim JW, et al. Identification of downstream genes of the mtor pathway that predict recurrence and progression in non-muscle invasive high-grade urothelial carcinoma of the bladder. J Korean Med Sci 2017;32:1327-36. [Crossref] [PubMed]


