
Comprehensive data analysis of genomics, epigenomics, and transcriptomics to identify specific biomolecular markers for prostate adenocarcinoma
Introduction
Prostate cancer is one of the most common malignancies in men worldwide. In the United States, the current cancer statistics indicate that emerging cases of prostate cancer may account for 21% of the 2020 cancer cases in men (1). Although the timely diagnostic strategies have been developed, approximately 20–35% of prostate adenocarcinoma (PRAD) cases inevitably progress into high-risk status with distal recurrence. Furthermore, around 10% of men diagnosed with PRAD die of their disease (2-4). The current strategy for local prostate cancer is laparoscopic prostatectomy, and the overall treatment for advanced patients is limited (5,6). Meanwhile, PRAD is characterized by tumor heterogeneity and frequently occurring metastasis, especially in the bone and bladder (7,8). Over the past years, the combination of enzalutamide and abiraterone, known as the androgen deprivation therapy (ADT), has been shown to be efficacious for most early-stage PRAD patients (9,10). However, some cases still develop into a stage of insensitivity and exhibit resistance to most ADT drugs (11,12). Other strategies, such as chemotherapy (docetaxel, oxaliplatin) and radiotherapy are used in a portion of metastatic and hormone-refractory cases treated by combined therapy (13). However, the overall benefits for patients are limited, with the side effects remain, including suppression of bone marrow, decreased appetite, and muscle aches, being problematic, (13-15). Therefore, novel therapeutic targets for prognostic prediction and improvements of PRAD need to be identified, along with better biomarkers for clarifying the potential mechanisms that lead to progression or distal metastasis of PRAD.
Recently, copy number variations (CNVs) or single-nucleotide variations (SNVs) that cause genomic variations have been reported to contribute to tumor development and recurrence (16-18). Many CNVs are reported to be closely associated with multiple pathological disorders, and it has been demonstrated that CNVs can directly regulate gene expression via altering messenger RNA (mRNA) levels or serial transcriptional regulation (19,20). Moreover, the association of DNA and histone methylation with the dysregulation of epigenomic instability in multiple malignancies has garnered considerable research attention (21). For instance, one study found that the tumor-suppressor genes in PRAD, including TP53, PTEN, and SPOP, were all hypermethylated and silenced across the CpG islands of gene promoter regions (22,23). Previous studies have already indicated that differential DNA methylation patterns are evident between normal and tumor samples, and between primary and metastatic cases (24,25). DNA methylation changes have been proven to correlate with differential risks of PRAD, and these associations with gene expression may facilitate the identification of PRAD the risk factors (26,27). Previous omics data analysis in specific cancers, like ovarian cancer or liver cancer, has already demonstrated that a wide range of epigenomic and genomic variations can influence tumor growth (28-30). Therefore, integrative analysis of based on high-throughput sequencing of multiomics data of large PRAD samples could help clarify the underlying risk factors of PRAD and provide novel insights into the relevance of aberrant gene levels in PRAD.
In the current study, we performed integrated multiomics data analysis (including genomics, methylomics, and transcriptomics) in 477 PRAD samples. Specifically, we determined the associations between mRNA levels and methylation variation (MET) or CNV to find specific METcor and CNVcor gene modules. Furthermore, we identified the specific subtypes of PRAD based on the screened METcor and CNVcor gene sets. Multivariate Cox regression analysis based on the 5 pivotal genes was conducted to construct the prognostic model for PRAD. A specific nomogram that incorporated risk scores and other parameters was also established to comprehensively illustrate the underlying abnormal variations identified in the omics data. We present the following article in accordance with the REMARK reporting checklist (available at https://dx.doi.org/10.21037/tau-21-576).
Methods
Data collection and preparation
We downloaded the mRNA expression data of PRAD samples and paired normal samples from The Cancer Genome Atlas (TCGA)-PRAD cohort (http://firebrowse.org/). We also acquired the single-nucleotide polymorphism (SNP)6 copy number segment and methylated data from the data portal. In total, 477 samples matched with complete SNP, CNV, and mRNA expression data were found, and the multiple omics data of the 477 samples were identified for the subsequent procedure. Those patients with incomplete sequencing or clinical data were excluded. For the CNV data processing, we filtered the area where the number of probes in the copy number segment was <5. For the methylation data, we deleted the sites with missing values. For the SNV data, we deleted the mutations in intron regions and silent variations. We used the GRCh38 release 22 (https://www.gencodegenes.org/human/release_22.html) to map the CNV region to corresponding genes. The samples with >0% absent loci were deleted to preprocess the MET data, and we used the K nearest neighbor (KNN) algorithm to impute the missing data (31). We maintained the probes in the transcription start site (TSS) from 2 kb upstream to 200 bp downstream, and mapped them to corresponding genes based on the GRCh38 release 22. We also screened the RNA-sequencing (RNA-seq) expression data and filtered the genes with low detecting levels [fragments per kilobase per million (FPKM) =0 in <0.5% of all samples]. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Identification of CNVcor- and METcor-associated gene sets
We calculated and assessed the associations between CNV and RNA-seq, and between MET and RNA-seq based on Pearson correlation coefficients. We used the Genomic Identification of Significant Targets in Cancer (GISTIC) method to detect the common CNV area in all samples, including the chromosome arm level CNV and the smallest common area across samples. The parameters of the GISTIC method in our study were as follows: Q ≤0.05 was set as the significance standard of change; when determining the peak interval, the confidence level was 0.95; and when analyzing the variation of the chromosome arm level, the region greater than the length of the chromosome arm (0.98) was considered the standard. The analysis was performed by the online analysis tool, GenePattern (Broad Institute; https://cloud.genepattern.org/gp/pages/index.jsf). We further used the “limma” package in R software to perform the differential gene analysis across the prostate cancer samples within the threshold standard of |log2-fold change (FC)| >1.0 and false discovery rate (FDR) <0.05. Furthermore, we calculated the Pearson correlation coefficients of CNV and RNA-seq, and MET and RNA-seq, respectively, and converted the correlation coefficient to z value according to the following formula: ln(1+r)/(1-r). In the correlation coefficient test, the genes with a P value <0.05 were considered to be the CNVcor and METcor gene sets. Finally, we used the R package “iCluster” (The R Foundation for Statistical Computing) to perform multigroup cluster analysis on CNVcor gene CNV data, METcor gene methylation data, and CNVcor and METcor gene mRNA data (expression matrix). After completing 50 iterations, the samples were divided into iC1, iC2, iC3, and iC4 clusters.
Screening of candidate genes and establishment of prognostic model
We conducted univariate Cox analysis to determine the prognostic significance of each candidate across the subgroups of the PRAD-cohort and regarded the genes with a P value <0.05 as significant. We then used multivariate Cox analysis to establish the prognostic model and determined the optimal threshold to divide the high-risk and low-risk PRAD samples using the “maxstat” package. Kaplan-Meier analysis was used to assess the predictive efficiency of the prognostic model. We then integrated the prognostic model with traditional clinical parameters to construct the nomogram model. A nomogram is also known as an alignment diagram, which integrates multiple predictive indicators and then uses scaled line segments according to a certain proportion drawn on the same plane to indicate the associations across the variables in the prediction model. Finally, concordance index (C-index) and receiver operating characteristic curve (ROC) analysis were used to compare the prediction accuracy of nomogram and other independent prognostic factors. All statistical tests were 2-tailed tests, and the statistical significance level of this study was set as P<0.05.
Calculation of immune scores for PRAD samples
Instead of just using one algorithm, TIMER2.0 (http://timer.cistrome.org/) provides more robust estimation of immune infiltration levels for The Cancer Genome Atlas (TCGA) or user-provided tumor profiles using six state-of-the-art algorithms. TIMER2.0 provides four modules for investigating the associations between immune infiltrates and genetic or clinical features,
Statistical analysis
The differentially expressed gene (DEG) analysis was conducted using the “limma” package in R software. Univariate and multivariate Cox analyses were conducted to screen the prognostic factors. The nomogram was constructed with the “rms” package, and Kaplan-Meier analysis was used to determine the prognostic difference between groups. All the statistical analysis was conducted in R studio (Version 3.6), and a P value <0.05 was considered statistically significant.
Results
Identification of CNVcor and METcor gene sets
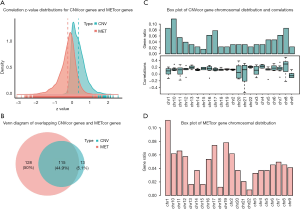
The clinical information of PRAD samples in our study is summarized in total online: https://cdn.amegroups.cn/static/public/tau-21-576-1.xls. We conducted the Pearson correlation analysis to calculate the associations among CNV, methylation, and mRNA of each gene. A list of 10,073 CNVcor genes and 9,841 METcor genes were confirmed with a significance level of P<0.01. Based on the z value distribution plot, we found that the correlation of CNVcor gene was significantly shifted to the right, while the correlation of METcor gene was significantly shifted to the left (Figure 1A). These data suggested that there is a negative association between gene expression and METcor genes and a positive association between gene expression and CNVcor genes. Due to the large amounts of genes in the 2 groups, we conducted differential analysis among tumor versus normal samples to screen out 598 DEGs using a cutoff of |log2-FC)| >1.0 and FDR <0.05. There were 371 genes associated with prognosis among the 598 DEGs (P<0.05). The Venn diagram also indicated the overlapping CNVcor genes and METcor genes; in Figure 1B, the green area represents the 128 CNVcor genes and the yellow area represents the 243 METcor genes The overlapping region contains 115 genes. The subsequent analysis also illustrated the genomic distributions of CNVcor and METcor genes, which indicated that CNVcor is more likely to appear on chromosome (chr)8, chr17, and chr10, while METcor is more likely to appear on chr1, chr19, and chr17 (Figure 1C,D, in total online: https://cdn.amegroups.cn/static/public/tau-21-576-2.xls).
Identification of molecular subtypes based on CNVcor and METcor genes
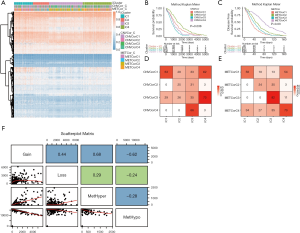
We used the nonnegative matrix factorization (NMF) algorithm to cluster the CNVcor and METcor gene sets from the 477 PRAD samples. Among the 50 interactions, the optimal number of clusters was determined when the classification number K was set from 2 to 5 according to cophenetic, dispersion, and silhouette. The NMF method in R studio was used to determine the average contour width of the matrix, and we set the minimal number for each subtype to be 10. We found that the optimal clustering number for CNVcor genes was 4 and the optimal clustering number for METcor genes was also 4 (Figure 2A,B). Interestingly, we observed a significant difference in the prognosis of both the CNVcor and METcor genes in the 4 subtypes through Kaplan-Meier analysis (P<0.05, Figure 2C,D). Finally, the subgroups clustered according to the CNVcor and METcor gene sets displayed a large amount of overlap (Figure 2E,F).
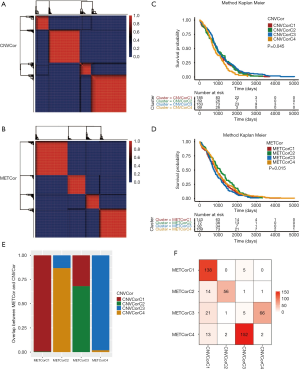
PRAD samples divided into 4 categories according to CNV, MET, and transcriptome data
According to the iCluster R package, we successfully divided the 477 PRAD samples into 4 subgroups based on the integrated cluster analysis of the CNV data of the CNVcor genes, methylated data of the METcor genes, and the mRNA data of the CNVcor and METcor genes. We determined that the minimal clustering number could not be less than 8, and we classified the samples into final subtypes, incorporating iC1 (92 samples), iC2 (79 samples), iC3 (165 samples), and iC4 (141 samples). We visualized the final clustering results of the PRAD cohort in a heatmap plot (Figure 3A). Meanwhile, we observed a significant difference in overall survival (OS) of the PRAD cohort across the 4 subtypes (Figure 3B,C). We also found a significant overlap between the iCluster gene clustering results and the gene results clustered by CNVcor and METcor (Figure 3D,E). In order to investigate the underlying relationship between CNV and MET variations, we defined the genes with a β value >0.3 as copy number amplification (CNA), and the genes with a β value <0.3 as CNV loss. Similarly, we considered the hypermethylation (MET-hyper) as a β value >0.8 and hypomethylation (MET-hypo) as a β value <0.2. We thus calculated the genes based on CNA, CNV loss, MET-hyper, and MET-hypo. We found that CNV gain and CNV loss were positively correlated (r=0.44) and that CNA and MET-hyper were strongly correlated (R2=0.68, P<0.0001; Figure 3F). CNV loss and MetHyper were also positively correlated (r=0.29; Figure 3F), whereas MetHypo and MetHyper were found to be negatively correlated (r=−0.28; Figure 3F).
Characterization of subtype features in PRAD and pivotal genes with altered CNV, MET, and mRNA
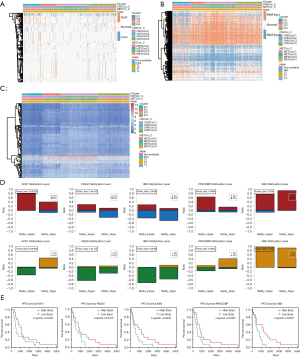
We integrated the CNV, MET, and mRNA data across the iC1, iC2, iC3, and iC4 subgroups and conducted differential analysis. According to the threshold defined above, each sample could be divided into CNA and CNV loss and similarly into MET-hyper and MET-hypo. A Fisher exact test was then used to identify genes with significant differences in CNV (944) and MET (1909) among iC1, iC2, iC3, and iC4 subtypes. The “limma” package was used to identify a list of 749 DEGs with the cutoff of |log2-FC)| >1.0 and FDR <0.05 across the 4 groups. The integrated heatmaps incorporating CNV, MET, and mRNA of DEGs are shown in Figure 4A,B,C. Of note, there were 6 genes with simultaneously altered CNA, MET, and mRNA, including IER3, AOX1, PRKCDBP, HLA-A, UBD, and FBLN5. Meanwhile, we specifically illustrated the CNV and mRNA of genes across clusters with respective prognostic significance (Figure 4D,E). Gene set enrichment analysis (GSEA) also revealed the potential pathways associated with these genes (Figure 5A,B,C).

Construction and assessment of the prognostic model for PRAD
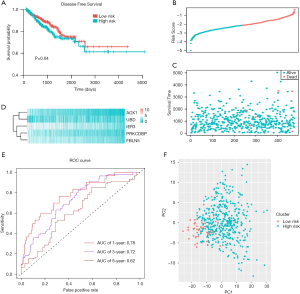
We conducted the multivariate Cox regression analysis based on the 5 genes (IER3, AOX1, PRKCDBP, UBD, and FBLN5) to construct the prognostic model for PRAD. We calculated the risk scores for each PRAD patient and obtained the optimal cutoff value based on the “maxstat” package. We thus could divide the whole PRAD-cohort into high-risk and low-risk groups according to the median score. We found that patients with high-risk scores suffered from worse progression-free survival (PFS) outcomes than did those with low-risk scores (Figure 6A). We ranked and drew the serial scores plot and the distribution diagram (Figure 6B,C,D). We further assessed the predictive accuracy of risk score for the PFS of PRAD, with the 1-, 3-, and 5-year area under the curve (AUC) being 0.78, 0.72, and 0.62, respectively (Figure 6E). We further conducted the principal component analysis (PCA) analysis and found that the risk scores could successfully classify the patients into 2 distinct groups (Figure 6F). To further determine whether the 5-gene risk score was independent of other clinical factors, we conducted univariate and multivariate regression to integrate these factors. The tumor-node-metastasis (TNM) stages and risk scores were both independent factors for PRAD (Figure 7A). We then constructed the nomogram for PRAD (Figure 7B). We calculated the specific nomogram-predicted probability of 1-, 3-, and 5-year PFS (Figure 7C). We also integrated the risk scores with N stage and found that the comprehensive model could effectively improve the predictive ability for PFS outcomes of PRAD, while the AUC of the nomogram reached up to 0.9 in predicting 1-, 3-, and 5-year PFS (Figure 7D).

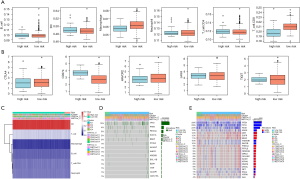
Integrated comparisons of SNP, CNV, and immune infiltrations between high-risk and low-risk PRAD patients
We utilized the TIMER platform to calculate the immune scores for each sample across groups. We observed that there were significant differences of cluster of differentiation (CD)4+ T cells and CD8+ T cells among the 4 subtypes (Figure 8A). Meanwhile, we also detected significant differential in the expression levels of common immune checkpoints (CTLA4, PDCD1, HAVCR2, LAG3, TIGIT) in high-risk and low-risk groups (Figure 8B). In addition, the programmed death-ligand 1 (PD-L1) expression levels were markedly upregulated in high-risk samples relative to those in low-risk samples. We thus speculated that there may exist differential immune infiltration patterns in the 2 risk groups, which might in turn impact the prognosis and survival of PRAD patients. We also used the Fisher exact test to identify the significant differential in genes between the 2 risk groups, among which there were 625 genes that showed differential mutation between the 2 groups. Among them, 522 genes showed differential CNV between the 2 risk groups. We screened and selected the top 10 significant genes and gene sets of partial interest. The CNV and mutated information of these genes were visualized in a heatmap (Figure 8C,D,E).
Discussion
Recently, high-throughput sequencing technology has become a novel strategy to identify potential markers and underlying mechanisms associated with tumor progression and prognosis (32-34). Meanwhile, large cancer databases, such as TCGA, International Cancer Genome Consortium (ICGC), and Gene Expression Omnibus (GEO), have been made available to researchers (35,36). These public sequencing databases can help researchers identify distinct molecular subtypes, potential cancer biomarkers, and candidate therapeutic inhibitors (37,38). For instance, Li et al. integrated the genomics, proteomics, and phosphoproteomics of 480 clinical tissues from 146 colon cancer patients and successfully screened a list of drug-targeted genes and a kinase-substrate network (39). Meanwhile, Kamoun et al. also successfully divided the eligible PRAD samples into 3 molecular subtypes with distinct genomic, transcriptomic, epigenomic, and clinical features (40). The emergence of this method has allowed for the integrative analysis of multiple omics that can comprehensively illustrate the genomic or transcriptional variations and identify the specific subgroups with distinct molecular features and tumor prognosis.
In the current study, we collected the multiomics data of 477 PRAD samples, including mRNA expression data, methylated data, CNV data, and corresponding clinical information. We first identified a list of 10,073 CNVcor genes and 9841 METcor genes that were confirmed with a significance level of P<0.01. We then implemented a NMF algorithm to cluster the CNVcor and METcor gene sets from the 477 PRAD samples. We found that the 4 classified subgroups based on METcor or CNVcor had significantly different prognoses. We further classified the samples into final subtypes, incorporating iC1 (92 samples), iC2 (79 samples), iC3 (165 samples), and iC4 (141 samples). We also identified 6 pivotal genes with simultaneously altered CNA, MET, and mRNA, including IER3, AOX1, PRKCDBP, HLA-A, UBD, and FBLN5. We used multivariate Cox regression analysis based on the 5 genes (IER3, AOX1, PRKCDBP, UBD, and FBLN5) to construct the prognostic model for PRAD. The high-risk and low-risk PRAD samples were accordingly classified based on the pivotal genes with distinct PFS outcomes. Finally, we confirmed that the 5-gene risk score could function as an independent factor compared with other clinical factors. The integrative risk nomogram proved to possess predictive ability for PRAD prognosis.
Previous studies have indicated that AXO1-associated metabolites have a high predictive significance for advanced bladder cancer and that AOX1 is epigenetically silenced during bladder cancer growth (41). Li et al. used the Cox proportional risk model to test a total of 126,633 SNPs to determine the potential relationships between PFS of PRAD and observed that SNP rs73055188 at the AOX1 locus correlated with prostate cancer-specific survival outcomes. Furthermore, AOX1 expression levels have also been associated with the biological recurrence of prostate cancer (42). Our analysis also supported AOX1 a risk factor in PRAD and samples, with those patients with a high expression of AOX1 experiencing worse PFS outcomes. One study using gene ontology (GO) analysis found AOX1 to be associated with biological processes, including cell migration, immune response, and cytoskeleton component. IER3 was also identified as an oncogene in our analysis, with PRAD patients with high IER3 levels having worse PFS outcomes. Similarly, Jordan et al. reported an activated inflammatory signaling network and indicated that the IL6-IER3 signaling axis contributed to chemoresistance and ovarian cancer recurrence (43).
We also constructed the prognostic model based on the 5 pivotal genes (IER3, AOX1, PRKCDBP, UBD, and FBLN5) and found that the established risk scores possessed high predictive accuracy for the PFS of PRAD. However, we only used the median risk score as the cutoff, but there may perhaps be a more appropriate algorithm to identify the optimal threshold. Second, we validated the predictive efficiency of risk scores in our larger prostate cancer cohort. Since the risk score still remained an independent prognostic factor compared with other clinical parameters, we constructed an integrative nomogram for PRAD patients. We calculated the specific nomogram-predicted probability of 1-, 3-, and 5-year PFS, and integrated the risk scores with N stage. This comprehensive model could efficiently improve the predictive ability for PFS outcomes of PRAD. However, we did not validate this model in other independent PRAD cohorts, and use of large samples is warranted to quantify the weight coefficients of each variate and the rational threshold for distinguishing high- and low-risk PRAD cases. Finally, we assessed the differential immune infiltration patterns in the high- and low-risk PRAD samples and observed the significant difference of CD8+ T cells. We observed that PD-L1 expression levels were markedly upregulated in high-risk samples. Thus, we speculated whether the dysregulation of the PDL-1–CD8+ T cell axis could lead to a worse prognosis in the high-risk groups. Antonarakis et al. recently published findings of the multicohort, open-label, phase II KEYNOTE-199 study, reporting good efficacy of pembrolizumab in treating refractory metastatic castration-resistant prostate cancer (44). Thus, whether immune checkpoint blockade (ICB) treatment is suitable for high-risk patients and whether specific regulations occur between the immunosuppressive condition and the 5 pivotal genes remain unclear.
In summary, our study integrated the multiomics data of genomics, epigenomics, and transcriptomics to uncover the potential pathogenic features of prostate cancer. We identified the pivotal METcor and CNVcor genes, and classified the 4 molecular subtypes of PRAD samples. We also constructed the specific prognostic model based on 5 hub genes and produced the corresponding nomogram. Finally, we integrated these data to develop a more accurate diagnosis and provide novel therapeutic targets for PRAD patients.
Acknowledgments
Funding: This study was supported by grants from the Natural Science Foundation of China (no. 82060533, 81860453, and 82072820) and Yunnan Provincial Science and Technology Department (no. 202001AY070001-055).
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at https://dx.doi.org/10.21037/tau-21-576
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/tau-21-576). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Goding Sauer A, et al. Colorectal cancer statistics, 2020. CA Cancer J Clin 2020;70:145-64. [Crossref] [PubMed]
- Litwin MS, Tan HJ. The Diagnosis and Treatment of Prostate Cancer: A Review. JAMA 2017;317:2532-42. [Crossref] [PubMed]
- Haberkorn U, Eder M, Kopka K, et al. New Strategies in Prostate Cancer: Prostate-Specific Membrane Antigen (PSMA) Ligands for Diagnosis and Therapy. Clin Cancer Res 2016;22:9-15. [Crossref] [PubMed]
- Chang AJ, Autio KA, Roach M, et al. High-risk prostate cancer-classification and therapy. Nat Rev Clin Oncol 2014;11:308-23. [Crossref] [PubMed]
- Petas A, Erickson A, Santti H, et al. Fast prostate retrieval in robot-assisted laparoscopic prostatectomy for next-generation biobanking. J Robot Surg 2020;14:271-4. [Crossref] [PubMed]
- Harper B, Klaassen Z, Wallis CJD. Local therapy in patients with metastatic prostate cancer: a new standard of care? Transl Cancer Res 2019;8:S592-4. [Crossref]
- Lange JM, Laviana AA, Penson DF, et al. Prostate cancer mortality and metastasis under different biopsy frequencies in North American active surveillance cohorts. Cancer 2020;126:583-92. [Crossref] [PubMed]
- Wu X, Lv D, Eftekhar M, et al. A new risk stratification system of prostate cancer to identify high-risk biochemical recurrence patients. Transl Androl Urol 2020;9:2572-86. [Crossref] [PubMed]
- Kim K, Watson PA, Lebdai S, et al. Androgen Deprivation Therapy Potentiates the Efficacy of Vascular Targeted Photodynamic Therapy of Prostate Cancer Xenografts. Clin Cancer Res 2018;24:2408-16. [Crossref] [PubMed]
- Greenberger BA, Zaorsky NG, Den RB. Comparison of Radical Prostatectomy Versus Radiation and Androgen Deprivation Therapy Strategies as Primary Treatment for High-risk Localized Prostate Cancer: A Systematic Review and Meta-analysis. Eur Urol Focus 2020;6:404-18. [Crossref] [PubMed]
- Obradovic AZ, Dallos MC, Zahurak ML, et al. T-Cell Infiltration and Adaptive Treg Resistance in Response to Androgen Deprivation With or Without Vaccination in Localized Prostate Cancer. Clin Cancer Res 2020;26:3182-92. [Crossref] [PubMed]
- Tousignant KD, Rockstroh A, Poad BLJ, et al. Therapy-induced lipid uptake and remodeling underpin ferroptosis hypersensitivity in prostate cancer. Cancer Metab 2020;8:11. [Crossref] [PubMed]
- Aparicio AM, Harzstark AL, Corn PG, et al. Platinum-based chemotherapy for variant castrate-resistant prostate cancer. Clin Cancer Res 2013;19:3621-30. [Crossref] [PubMed]
- Chi KN, Agarwal N, Bjartell A, et al. Apalutamide for Metastatic, Castration-Sensitive Prostate Cancer. N Engl J Med 2019;381:13-24. [Crossref] [PubMed]
- Davis ID, Martin AJ, Stockler MR, et al. Enzalutamide with Standard First-Line Therapy in Metastatic Prostate Cancer. N Engl J Med 2019;381:121-31. [Crossref] [PubMed]
- Verhaak RGW, Bafna V, Mischel PS. Extrachromosomal oncogene amplification in tumour pathogenesis and evolution. Nat Rev Cancer 2019;19:283-8. [Crossref] [PubMed]
- Kutilin DS, Airapetova TG, Anistratov PA, et al. Copy Number Variation in Tumor Cells and Extracellular DNA in Patients with Lung Adenocarcinoma. Bull Exp Biol Med 2019;167:771-8. [Crossref] [PubMed]
- Wu S, Li G, Zhao X, et al. High-level gain of mesenchymal-epithelial transition factor (MET) copy number using next-generation sequencing as a predictive biomarker for MET inhibitor efficacy. Ann Transl Med 2020;8:685. [Crossref] [PubMed]
- O'Hara AJ, Le Gallo M, Rudd ML, et al. High-resolution copy number analysis of clear cell endometrial carcinoma. Cancer Genet 2020;240:5-14. [Crossref] [PubMed]
- Marcon J, DiNatale RG, Sanchez A, et al. Comprehensive Genomic Analysis of Translocation Renal Cell Carcinoma Reveals Copy-Number Variations as Drivers of Disease Progression. Clin Cancer Res 2020;26:3629-40. [Crossref] [PubMed]
- Clarke TL, Tang R, Chakraborty D, et al. Histone Lysine Methylation Dynamics Control DNA Copy-Number Amplification. Cancer Discov 2020;10:306-25. [Crossref] [PubMed]
- Chen S, Wang Q, Yu H, et al. Mutant p53 drives clonal hematopoiesis through modulating epigenetic pathway. Nat Commun 2019;10:5649. [Crossref] [PubMed]
- Zhang J, Lee YR, Dang F, et al. PTEN Methylation by NSD2 Controls Cellular Sensitivity to DNA Damage. Cancer Discov 2019;9:1306-23. [Crossref] [PubMed]
- Gkountela S, Castro-Giner F, Szczerba BM, et al. Circulating Tumor Cell Clustering Shapes DNA Methylation to Enable Metastasis Seeding. Cell 2019;176:98-112.e14. [Crossref] [PubMed]
- Rodger EJ, Chatterjee A, Stockwell PA, et al. Characterisation of DNA methylation changes in EBF3 and TBC1D16 associated with tumour progression and metastasis in multiple cancer types. Clin Epigenetics 2019;11:114. [Crossref] [PubMed]
- Pellacani D, Droop AP, Frame FM, et al. Phenotype-independent DNA methylation changes in prostate cancer. Br J Cancer 2018;119:1133-43. [Crossref] [PubMed]
- Patel PG, Wessel T, Kawashima A, et al. A three-gene DNA methylation biomarker accurately classifies early stage prostate cancer. Prostate 2019;79:1705-14. [Crossref] [PubMed]
- Chaudhary K, Poirion OB, Lu L, et al. Deep Learning-Based Multi-Omics Integration Robustly Predicts Survival in Liver Cancer. Clin Cancer Res 2018;24:1248-59. [Crossref] [PubMed]
- Dimitrakopoulos C, Hindupur SK, Häfliger L, et al. Network-based integration of multi-omics data for prioritizing cancer genes. Bioinformatics 2018;34:2441-8. [Crossref] [PubMed]
- Rappoport N, Shamir R. Multi-omic and multi-view clustering algorithms: review and cancer benchmark. Nucleic Acids Res 2018;46:10546-62. [Crossref] [PubMed]
- Arian R, Hariri A, Mehridehnavi A, et al. Protein kinase inhibitors' classification using K-Nearest neighbor algorithm. Comput Biol Chem 2020;86:107269 [Crossref] [PubMed]
- Callahan BJ, Wong J, Heiner C, et al. High-throughput amplicon sequencing of the full-length 16S rRNA gene with single-nucleotide resolution. Nucleic Acids Res 2019;47:e103 [Crossref] [PubMed]
- Zhang X, Li T, Liu F, et al. Comparative Analysis of Droplet-Based Ultra-High-Throughput Single-Cell RNA-Seq Systems. Mol Cell 2019;73:130-42.e5. [Crossref] [PubMed]
- Cheng YH, Chen YC, Lin E, et al. Hydro-Seq enables contamination-free high-throughput single-cell RNA-sequencing for circulating tumor cells. Nat Commun 2019;10:2163. [Crossref] [PubMed]
- ICGC/TCGA Pan-Cancer Analysis of Whole Genomes Consortium. Pan-cancer analysis of whole genomes. Nature 2020;578:82-93. [Crossref] [PubMed]
- Gerstung M, Jolly C, Leshchiner I, et al. The evolutionary history of 2,658 cancers. Nature 2020;578:122-8. [Crossref] [PubMed]
- Tan TZ, Rouanne M, Tan KT, et al. Molecular Subtypes of Urothelial Bladder Cancer: Results from a Meta-cohort Analysis of 2411 Tumors. Eur Urol 2019;75:423-32. [Crossref] [PubMed]
- Laddha SV, da Silva EM, Robzyk K, et al. Integrative Genomic Characterization Identifies Molecular Subtypes of Lung Carcinoids. Cancer Res 2019;79:4339-47. [Crossref] [PubMed]
- Li C, Sun YD, Yu GY, et al. Integrated Omics of Metastatic Colorectal Cancer. Cancer Cell 2020;38:734-47.e9. [Crossref] [PubMed]
- Kamoun A, Cancel-Tassin G, Fromont G, et al. Comprehensive molecular classification of localized prostate adenocarcinoma reveals a tumour subtype predictive of non-aggressive disease. Ann Oncol 2018;29:1814-21. [Crossref] [PubMed]
- Vantaku V, Putluri V, Bader DA, et al. Correction: Epigenetic loss of AOX1 expression via EZH2 leads to metabolic deregulations and promotes bladder cancer progression. Oncogene 2020;39:6387-92. [Crossref] [PubMed]
- Li W, Middha M, Bicak M, et al. Genome-wide Scan Identifies Role for AOX1 in Prostate Cancer Survival. Eur Urol 2018;74:710-9. [Crossref] [PubMed]
- Jordan KR, Sikora MJ, Slansky JE, et al. The Capacity of the Ovarian Cancer Tumor Microenvironment to Integrate Inflammation Signaling Conveys a Shorter Disease-free Interval. Clin Cancer Res 2020;26:6362-73. [Crossref] [PubMed]
- Antonarakis ES, Piulats JM, Gross-Goupil M, et al. Pembrolizumab for Treatment-Refractory Metastatic Castration-Resistant Prostate Cancer: Multicohort, Open-Label Phase II KEYNOTE-199 Study. J Clin Oncol 2020;38:395-405. [Crossref] [PubMed]
(English Language Editor: J. Jones)


