Identification of autophagy-related long non-coding RNA prognostic and immune signature for clear cell renal cell carcinoma
Introduction
Renal cell carcinoma (RCC) occupied approximately 3% of all cancers, with an annual increase of 2% incidence, leading to more than 400,000 new cases and 175,098 deaths worldwide in 2018 (1). Kidney renal clear cell carcinoma (KIRC) is a rather aggressive subtype representing about 85% of metastatic RCC cases and 67% of all stage RCC (2). Benefiting from the development of modern surgery and targeted drug research, most early-stage KIRC patients have a relatively high 5-year survival rate. However, approximately 25% to 30% of cases accompany metastases at diagnosis and 20% to 30% of patients show relapse after undergoing surgical management for local RCC (3), making the 5-year survival rate drop to only 23% (4). Lacking of reliable and stable prognostic markers bears responsibility for that. Presently, the clinical prognosis of KIRC patients is predicted by multidimensional factors, including clinical, anatomical, molecular factors, and histological with non-reliable guideline-approved biomarkers (5). For molecular factors, CAIX, PTEN, and CXCR4, etc. have been investigated one after another, however none of which has yet improved the current prognostic systems (6).
Protein-coding RNAs account for only 1–2% of the human genome and more than 90% of RNAs are thought to carry non-protein-coding information with other functions, modifying or regulating, etc. (7). During the past decade, the role of lncRNAs in RCC has been clearly highlighted. HOTAIR which is elevated in RCC cells is one of the few well-described lncRNAs that could be further investigated as a reliable molecular marker and promising target for RCC patients (8). Likewise, H19 is considered as an oncogene in RCC which plays an essential role in the epithelial-to-mesenchymal transition (EMT) process by mediating the function of EZH2, E-cadherin, and β-catenin (9). In addition to tumorigenesis and progression, lncRNA was also demonstrated to regulate drug resistance in RCC. SRLR (10), ARSR (11), and NEAT1 (12) were believed to promote drug resistance. In contrast, GAS5 was known to enhance RCC cell’s sensitivity to sorafenib via the GAS5/miR-21/SOX5 axis (13).
Since first reported 40 years ago, autophagy is involved in the pathologic process of various diseases, including cancer (14), cardiovascular disease (15), infection, and immune deficiency (16). Recently, the association between autophagy and RCC has been studied in signaling pathways and drug sensibility. For example, autophagy in RCC cells can be inhibited by the activation of the PI3K/AKT/mTOR axis, accompanied by protein translation and cell proliferation (17). Besides, sunitinib was demonstrated to cause autophagy in RCC cells by inhibiting AKT/mTOR signaling pathway (18).
In this work, we identified 17 autophagy-related lncRNAs through bioinformatics methods. Following prognosis analyses revealed that these lncRNAs were significantly associated with KIRC patients’ survival. Finally, immune-related pathways were dramatically enriched in high-risk groups according to GSEA results. Further CIBERSORT analysis revealed that these lncRNAs have significant impact on ccRCC tumor immune cells’ infiltration. We present the following article in accordance with the MDAR reporting checklist (available at https://dx.doi.org/10.21037/tau-21-278).
Methods
Workflow
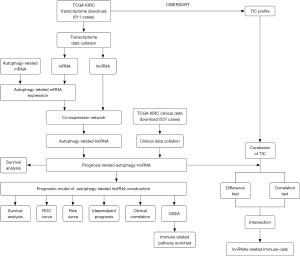
The steps we used in this work to screen autophagy-related lncRNAs and establish a prognosis model for KIRC were showed in Figure 1.
Data collection and pre-processing
We downloaded transcriptome profiling of KIRC from TCGA (https://portal.gdc.cancer.gov/) and selected HTseq-FPKM. A total of 611 cases were included in our subsequent analyses. We next separated RNA to mRNA and lncRNA using human.gtf downloaded from Ensembl database (http://asia.ensembl.org/index.html). Autophagy related mRNA gene list was downloaded from Human Autophagy Database (HADb, http://www.autophagy.lu/index.html). By merging this gene list and mRNA expression matrix, we got the autophagy-related mRNA gene expression. Clinical information was also obtained from TCGA and 537 cases were included for prognostic analysis.
Co-expression network
To screen autophagy-related lncRNA, we constructed mRNA-lncRNA co-expression network by using the R language. The correlation coefficient |R|>0.3 and P value <0.0001 were considered significantly correlated. The co-expression network between autophagy-related mRNA and lncRNA was mapped in Cytoscape (v3.7.2).
Risk score calculation
First, clinical data and autophagy-related lncRNA expression were combined to get the relationship between expression and survival time. We then performed univariate and multivariate Cox regression analyses to evaluate the prognostic value of these lncRNAs. The lncRNAs with a P value <1E-6 by univariate analysis were further enrolled to conduct multivariate Cox regression analysis to assess the risk score. The Akaike information criterion (AIC) values were used to optimize the Cox model and the lncRNAs with the lowest AIC were retained in the final signature. The risk score formula was as followed: Risk score = ∑lncRNAiexp×lncRNAicoef (lncRNAiexp indicates the expression of every single lncRNA and the lncRNAicoef was calculated using a multivariable Cox proportional hazards model). All patients were divided into high or low-risk group basing on the median risk score. Sankey diagram was built with the “ggalluvial” package according to the HR (hazard ratio) value of multivariate Cox proportional hazards regression analyses (HR >1 was considered as risk gene, HR<1 was considered as protect gene.).
Prognosis model construction
Given the prognosis related lncRNAs, we subsequently constructed a prognosis model to assess the prognostic value of these lncRNAs for KIRC patients. The overall survival (OS) curves were plotted with “survival” package basing on the expression of lncRNAs or risk score. The independence of risk signature for prognosis was further evaluated by univariate and multivariate Cox proportional hazards regression analyses. For a more intuitive understanding of the relevance between lncRNAs expression and patients’ survival state, we drew the risk score curve, survival time, and heatmap. The heatmap was drawn with “pheatmap” packages. To evaluate the signature’s sensitivity and specificity, the clinical variables dependent receiver operating characteristic (ROC) analyses were performed with “survivalROC” package. Additionally, to explore the correlation between clinical features and risk score, we excluded patients with deficient information and 246 patients were retained for analysis.
Gene set enrichment analysis (GSEA)
611 KIRC samples in TCGA were divided into two groups (high risk and low risk) based on the median of risk scores in lncRNA signature. We conducted GSEA between the two groups to identify the significantly altered Gene Ontology (GO) pathways by using GSEA software (v4.0.3). C5.all.v6.1.symbols.gmt was used as the gene set (19,20). The P and FDR q values were obtained from 1,000 permutations and P<0.05 was considered statistically significant.
Immune cell infiltration analysis
CIBERSORT analysis was used to assess the relevance of 22 tumor-infiltrating immune cells in ccRCC tumor tissues and autophagy related lncRNAs expression. For difference test, samples were divided into a low expression group (lncRNAlow) and a high expression group (lncRNAhigh) based on the median expression, respectively. The difference of immune cells infiltrating level between these groups was compared with Wilcoxon rank-sum test, and P value <0.05 was considered significant. For correlation test, spearman test was utilized. Afterward, intersection of the two tests results was regarded as lncRNA related immune cells in ccRCC tumor tissues.
Statistical analysis
Almost all analyses were performed with R software (4.0.0) or Perl (5.32.0). The Student’s t-test was used to compare the differences between two or three groups. P value <0.05 was considered statistically significant.
Ethics
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and national research committee and with the Helsinki Declaration (as revised in 2013). Written informed consent was obtained from the patient.
Results
17 lncRNAs were identified as autophagy-related
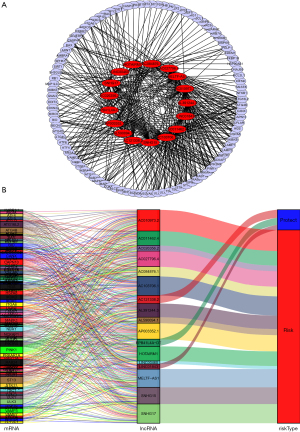
A total of 14,142 lncRNAs were distinguished from the transcriptome matrix downloaded from the TCGA database. 258 autophagy-related mRNAs were obtained from the HADb database. The co-expression network was constructed to identify autophagy-related lncRNAs. Finally, 17 lncRNAs and 99 autophagy-related mRNA were screened out and considered as significant correlation (|R|>0.3, P value <0.0001). We then used these RNAs as nodes to draw a network (Figure 2A). Furthermore, we created this image (Figure 2B)—a Sankey diagram—by plotting the connection within each node and connecting them to risk type which based on the HR value of multivariate Cox proportional hazards regression analyses (HR >1 was considered as risk gene, HR <1 was considered as protecting gene). Three of them were defined as protect genes (AC121338.2, EPB41L4A-DT, LINC01843) and others were all risk genes (AL391244.3, AC011462.4, AC103706.1, SNHG15, AL590094.1, AP003352.1, AC026356.2, SNHG17, LINC00460, HOTAIRM1, AC084876.1, AC027796.4, MELTF-AS1, AC010973.2). The univariate Cox regression for each individual lncRNAs was provided in Table S1.
Survival analyses of these 17 autophagy-related lncRNAs
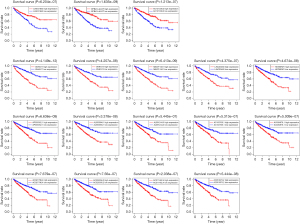
Given the 17 autophagy-related lncRNAs, we next explored their impact of expression level on OS (Figure 3). For three protective autophagy-related lncRNAs, they showed a significant tendency that higher expression predicting better survival rate. In contrast, for 14 risky autophagy-related lncRNAs, higher expression may lead to poorer survival rates. The survival analysis results were consistent with the risk type classified by multivariate Cox proportional hazards regression analyses.
Risk signature assessment
After performing survival analyses of these autophagy-related lncRNAs, we next constructed the prognosis assessment of risk signature. First, we divided patients into high-risk group and low-risk group to compare their survival difference. As shown in Figure 4A, the high-risk group displayed a lower survival rate than the low-risk group. Next, we plotted the expression patterns of the 17 autophagy-related lncRNAs, survival status dot plot, and risk score curve to visualize the association between these variables (Figure 4B). The heatmap showed that 3 protective autophagy-related lncRNAs (LINC01843, EPB41L4A-DT, and AC121338.2) were down-regulated in high-risk group whereas other lncRNAs were overexpressed. Additionally, the survival status dot plot revealed that dead cases were almost concentrated in the high-risk group and more dead cases appeared with the increase of risk score.
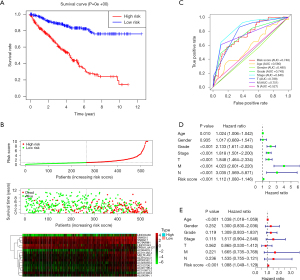
In the ROC analysis (Figure 4C), the prognostic model was evaluated using the area under the curve (AUC). The AUC for the risk score signature was 0.748, higher than age (0.580), gender (0.480), grade (0.743) and N stage (0.527, lymph node metastasis), but lower than stage (0.846), T stage (0.788, the extent of the primary tumor) as well as M stage (0.751, distant metastasis). The results suggested that the prognostic value of risk signature was better than age, gender, grade, and N stage.
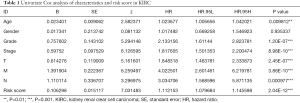
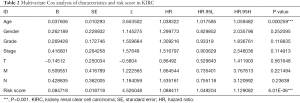
For the independent prognosis analysis, age, gender, grade, stage, TNM system, and risk score were included in the independent prognostic factors. Univariate (Figure 4D) and multivariate (Figure 4E) analyses suggested that the risk signature was an independent predictive factor for KIRC patients (P<0.001, HRs were 1.112 and 1.088, respectively). The details of analysis were shown in Table 1 and Table 2.
Full table
Full table
Clinical value of the risk signature for KIRC patients
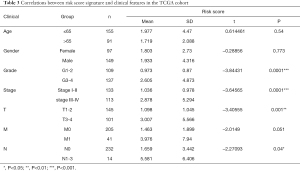
Subsequently, we evaluated the clinical value of the risk signature regarding age, gender, grade, stage, and TNM. As showed in Table 3, the risk score tended to elevate in higher grade (grade 3-4, P=0.0001), advanced stage (stage III-IV, P=0.0001), higher T stage (P=0.001) and N stage (P=0.04). These results suggested that the risk signature might be associated with the progression of KIRC.
Full table
Identified autophagy-related lncRNAs may function in tumor progression via Immunomodulation
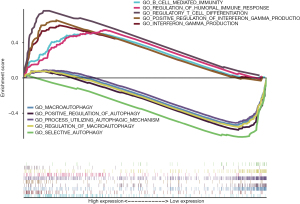
To explore the potential mechanism of these 17 autophagy-related lncRNAs’ effect on tumor progression, we conducted Gene Ontology (GO) functional annotation with GSEA. Interestingly, immune-related pathways were markedly enriched in the high-risk group, including regulatory T cell differentiation, positive regulation of interferon gamma production, interferon gamma production, regulation of humoral immune response, B cell mediated immunity. In low-risk group, autophagy-related pathways were significantly enriched, such as selective autophagy, regulation of macroautophagy, macroautophagy, process utilizing autophagic mechanism, and positive regulation of autophagy (Figure 5). The enrichment results were summarized in Table 4.
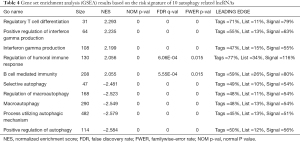
Full table
Identification of lncRNAs related tumor-infiltrating immune cells in ccRCC tumor tissues
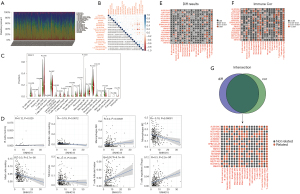
Given the pathway enriched in immune regulation, we next conducted analyses of the relevance of tumor immune cells infiltrating level and lncRNAs expression via CIBERSORT test. Immune cells infiltrating percent in every sample was showed in Figure 6A, and the correlation between these 22 immune cells was showed in Figure 6B. Difference test of SNHG15 (Figure 6C) revealed that B cells naive, B cells memory, T cells CD8, T cells CD4 memory resting, T cells follicular helper, T cells regulatory (Tregs), monocytes, macrophages M0, macrophages M2, dendritic cells resting and mast cells resting were significantly different between SNHG15high group and SNHG15low group. Meanwhile, B cells memory, T cells follicular helper, T cells regulatory (Tregs), monocytes, macrophages M0, macrophages M2, dendritic cells resting, and mast cells resting were identified SNHG15 correlative immune cells (Figure 6D). The major immune cells infiltrating results of difference test and correlation test for all the 17 autophagy-related lncRNAs were showed in Figure 6E,F. The intersection of the two tests was showed in Figure 6G, which made a landscape of autophagy-related lncRNAs’ correlative tumor-infiltrating immune cells in ccRCC tumor tissues.
Discussion
LncRNA/miRNA/mRNA axis is a promising target for tumor treatment (21). Given the important role of lncRNAs and autophagy in KIRC tumorigenesis, progression, and drug resistance, we performed this work to screen autophagy-related lncRNAs and assess their prognostic value for KIRC patients.
Among our identification of lncRNAs, several have been demonstrated to function in KIRC or other malignant tumors. Small nucleolar RNA host gene 15 (SNHG15) was upregulated in KIRC, knockdown of it inhibited KIRC cell proliferation, invasion, and migration (22). The EMT process induced by nuclear factor-κB signaling pathway may be the potential mechanism (22). Small nucleolar RNA host gene 17 (SNHG17) was reported to accelerate cell proliferation and invasion in castration-resistant prostate cancer (CRPC) by targeting the miR-144/CD51 axis (23). Furthermore, as lncRNAs that encode small nucleolar RNAs (snoRNAs), the prognostic value of SNHG17 and SNHG15 in KIRC patients has been investigated, and DNA hypomethylation might play a key role in elevated SNHG15 transcription in KIRC (24). Recently, AC026356.2 and MELTF-AS1 were identified as immune-related lncRNA and showed a significant relationship with KIRC prognosis (25). Zhang et al. established a lncRNA prognostic model based on LINC00460, MIAT, and LINC00443 from a competitive endogenous RNA regulatory network constructed in KIRC (26). The function of HOTAIRM1 in KIRC was reported by Hamilton et al. (27). It suggested that HOTAIRM1 served a crucial role in kidney differentiation and suppressed angiogenic pathways induced by HIF1. Noteworthily, HOTAIRM1 is one of the few reported lncRNAs whose function is involved in the autophagy pathway. The degradation of PML-RARA oncoprotein and differentiation of myeloid cell is regulated by HOTAIRM1 via enhancing the autophagy pathway (28).
In recent years, some studies constructed prognostic signatures for ccRCC based on autophagy-related genes (ARGs) to predict the prognosis of ccRCC. In Chen’s study (29), 11 ARGS were screened out and merged to construct a risk model. The area under the curve value of ROC for OS is 0.738. A seven-gene prognostic risk signature was reported in Yang’s study with an area under the curve value of 0.71 (30). In this study, we identified 17 lncRNAs based on their expression correlation with ARGs. The area under the curve value of ROC for OS is 0.748, slightly better than the reported two models. The fields of lncRNAs and autophagy are developing rapidly, and the regulatory role of lncRNA on autophagy in cancer also raised more attention in recent years. Many studies suggest that lncRNAs activate or inhibit autophagy by regulating autophagy-related genes and pathways, leading to promote or suppress tumor progression, depending on tumor microenvironment or cell environment pressure (31). Hence, there are four functional forms of cross regulation between lncRNAs and autophagy in cancer, lncRNAs promoting cancer by activating autophagy or inhibiting autophagy, lncRNAs suppressing cancer by activating autophagy or inhibiting autophagy (31). In renal cancer, some autophagy-related lncRNAs have already been well studied. HOTAIR was reported to negatively target miR-17-5p to activate cell autophagy which mediated by Beclin1, resulting in sunitinib resistance in renal cancer cells (32). In the study of Su et al. (33), 3-MA (an autophagy inhibitor) could reverse the inhibition of RCC cell proliferation, migration, and invasion induced by HOTTIP silencing. Further research found that the modification of HOTTIP affected RCC cell autophagy through the PI3K/Akt/Atg13 signaling pathway. In addition, RCC chemoresistance was known to be enhanced by lncRNA KIF9-AS1 which regulates autophagy signaling via miRNA-497-5p (34). In summary, the role of autophagy regulated by lncRNAs in RCC is not only tumor progression, but also drug resistance.
Recent studies have shown that immune responses for tumor tissues were significantly controlled by autophagy via modulating the functions of immune cells. In RCC, chloroquine (an autophagy inhibitor) was reported to improve high-dose IL-2 mediated anti-tumor immunity by enhancing T-cells, dendritic cells and NK cells (35). Besides, the anti-RCC effect of sunitinib is enhanced by chloroquine via inhibiting the autophagy which induced by sunitinib (36). The autophagy sensor ITPR1 was found to protect renal carcinoma cells from NK-mediated killing (37). In vitro, THZ1 acted synergistically with temsirolimus by inhibiting autophagy and induced cell cycle arrest and apoptosis in RCC cells (38).
In our current work, 17 autophagy-related lncRNAs were firstly identified in KIRC. The prognosis model was established, and the results indicated that the risk signature is a promising predictive indicator for patients with KIRC. Furthermore, to extract biological meaning from the identified differentially risk score, GSEA analysis was performed and immune-related GO terms were significantly enriched. These results declared that for high-risk patients, the immune response may be activated by the autophagy-related lncRNAs-miRNA-mRNA axis. Further studies should be performed to discover the miRNAs which connect the lncRNAs and mRNA to transmit signals and affect the immune response, transforming our identified autophagy-related lncRNAs into potential therapeutic targets for KIRC patients finally.
Conclusions
We successfully identified 17 autophagy-related lncRNAs and constructed a risk signature correlated with KIRC prognosis in the TCGA cohort. The results revealed that the signature is a potent and independent prognostic indicator for KIRC patients. Subsequent GSEA analysis showed that the immune-related pathways were distinctly enriched in high-risk group, which reminded us to make a hypothesis that these autophagy-related lncRNAs may regulate immune response by affecting autophagy process in RCC cells. Further analysis brought insight into the correlation between lncRNAs and tumor-infiltrating immune cells. Future studies concentrating on our signature may disclose novel therapeutic targets for KIRC patients.
Acknowledgments
We would like to thank editage (
Funding: This study was supported by the Six Talent Peak Project of High-level Talents in Jiangsu Province (WSW-017); 333 High-level Talents Training Project in Jiangsu Province, Professional from Six-Pronged Top-Talent Program (LGY2018053); Qing Lan Project of Jiangsu University (JX2161015100); The Fifth Batch of Outstanding Young and Middle-aged Teachers Support Program of Nanjing Medical University; A Project Funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions (JX10231802); Postgraduate Research & Practice Innovation Program of Jiangsu Province (KYCX19_1159) and International Exchange and Cooperation Program for Postgraduates of Nanjing Medical University.
Footnote
Reporting Checklist: The authors have completed the MDAR checklist. Available at https://dx.doi.org/10.21037/tau-21-278
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/tau-21-278). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and national research committee and with the Helsinki Declaration (as revised in 2013). Written informed consent was obtained from the patient.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Ferlay J, Colombet M, Soerjomataram I, et al. Cancer incidence and mortality patterns in Europe: Estimates for 40 countries and 25 major cancers in 2018. Eur J Cancer 2018;103:356-87. [Crossref] [PubMed]
- Kroeger N, Xie W, Lee JL, et al. Metastatic non-clear cell renal cell carcinoma treated with targeted therapy agents: characterization of survival outcome and application of the International mRCC Database Consortium criteria. Cancer 2013;119:2999-3006. [Crossref] [PubMed]
- Ljungberg B, Campbell SC, Choi HY, et al. The epidemiology of renal cell carcinoma. Eur Urol 2011;60:615-21. [Crossref] [PubMed]
- Ljungberg B, Bensalah K, Canfield S, et al. EAU guidelines on renal cell carcinoma: 2014 update. Eur Urol 2015;67:913-24. [Crossref] [PubMed]
- Ljungberg B, Albiges L, Abu-Ghanem Y, et al. European Association of Urology Guidelines on Renal Cell Carcinoma: The 2019 Update. Eur Urol 2019;75:799-810. [Crossref] [PubMed]
- Inamura K. Renal Cell Tumors: Understanding Their Molecular Pathological Epidemiology and the 2016 WHO Classification. Int J Mol Sci 2017;18:2195. [Crossref] [PubMed]
- Dinger ME, Pang KC, Mercer TR, et al. Differentiating protein-coding and noncoding RNA: challenges and ambiguities. PLoS Comput Biol 2008;4:e1000176 [Crossref] [PubMed]
- Wu Y, Liu J, Zheng Y, et al. Suppressed expression of long non-coding RNA HOTAIR inhibits proliferation and tumourigenicity of renal carcinoma cells. Tumour Biol 2014;35:11887-94. [Crossref] [PubMed]
- Raveh E, Matouk IJ, Gilon M, et al. The H19 Long non-coding RNA in cancer initiation, progression and metastasis - a proposed unifying theory. Mol Cancer 2015;14:184. [Crossref] [PubMed]
- Xu Z, Yang F, Wei D, et al. Long noncoding RNA-SRLR elicits intrinsic sorafenib resistance via evoking IL-6/STAT3 axis in renal cell carcinoma. Oncogene 2017;36:1965-77. [Crossref] [PubMed]
- Qu L, Ding J, Chen C, et al. Exosome-Transmitted lncARSR Promotes Sunitinib Resistance in Renal Cancer by Acting as a Competing Endogenous RNA. Cancer Cell 2016;29:653-68. [Crossref] [PubMed]
- Shin VY, Chen J, Cheuk IW, et al. Long non-coding RNA NEAT1 confers oncogenic role in triple-negative breast cancer through modulating chemoresistance and cancer stemness. Cell Death Dis 2019;10:270. [Crossref] [PubMed]
- Liu L, Pang X, Shang W, et al. Long non-coding RNA GAS5 sensitizes renal cell carcinoma to sorafenib via miR-21/SOX5 pathway. Cell Cycle 2019;18:257-63. [Crossref] [PubMed]
- Dikic I, Johansen T, Kirkin V. Selective autophagy in cancer development and therapy. Cancer Res 2010;70:3431-4. [Crossref] [PubMed]
- Liao X, Sluimer JC, Wang Y, et al. Macrophage autophagy plays a protective role in advanced atherosclerosis. Cell Metab 2012;15:545-53. [Crossref] [PubMed]
- Kim JJ, Lee HM, Shin DM, et al. Host cell autophagy activated by antibiotics is required for their effective antimycobacterial drug action. Cell Host Microbe 2012;11:457-68. [Crossref] [PubMed]
- Arico S, Petiot A, Bauvy C, et al. The tumor suppressor PTEN positively regulates macroautophagy by inhibiting the phosphatidylinositol 3-kinase/protein kinase B pathway. J Biol Chem 2001;276:35243-6. [Crossref] [PubMed]
- Cao P, Jiang XJ, Xi ZJ. Sunitinib induces autophagy via suppressing Akt/mTOR pathway in renal cell carcinoma. Beijing Da Xue Xue Bao Yi Xue Ban 2016;48:584-9.
- Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545-50. [Crossref] [PubMed]
- Mootha VK, Lindgren CM, Eriksson KF, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet 2003;34:267-73. [Crossref] [PubMed]
- Barth DA, Juracek J, Slaby O, et al. lncRNA and Mechanisms of Drug Resistance in Cancers of the Genitourinary System. Cancers (Basel) 2020;12:2148. [Crossref] [PubMed]
- Du Y, Kong C, Zhu Y, et al. Knockdown of SNHG15 suppresses renal cell carcinoma proliferation and EMT by regulating the NF-κB signaling pathway. Int J Oncol 2018;53:384-94. [Crossref] [PubMed]
- Bai M, Lei Y, Wang M, et al. Long Non-coding RNA SNHG17 Promotes Cell Proliferation and Invasion in Castration-Resistant Prostate Cancer by Targeting the miR-144/CD51 Axis. Front Genet 2020;11:274. [Crossref] [PubMed]
- Yang W, Zhang K, Li L, et al. Discovery and validation of the prognostic value of the lncRNAs encoding snoRNAs in patients with clear cell renal cell carcinoma. Aging (Albany NY) 2020;12:4424-44. [Crossref] [PubMed]
- Jiang Y, Gou X, Wei Z, et al. Bioinformatics profiling integrating a three immune-related long non-coding RNA signature as a prognostic model for clear cell renal cell carcinoma. Cancer Cell Int 2020;20:166. [Crossref] [PubMed]
- Zhang D, Zeng S, Hu X. Identification of a three-long noncoding RNA prognostic model involved competitive endogenous RNA in kidney renal clear cell carcinoma. Cancer Cell Int 2020;20:319. [Crossref] [PubMed]
- Hamilton MJ, Young M, Jang K, et al. HOTAIRM1 lncRNA is downregulated in clear cell renal cell carcinoma and inhibits the hypoxia pathway. Cancer Lett 2020;472:50-8. [Crossref] [PubMed]
- Chen ZH, Wang WT, Huang W, et al. The lncRNA HOTAIRM1 regulates the degradation of PML-RARA oncoprotein and myeloid cell differentiation by enhancing the autophagy pathway. Cell Death Differ 2017;24:212-24. [Crossref] [PubMed]
- Chen M, Zhang S, Nie Z, et al. Identification of an Autophagy-Related Prognostic Signature for Clear Cell Renal Cell Carcinoma. Front Oncol 2020;10:873. [Crossref] [PubMed]
- Yang H, Han M, Li H. Construction and Validation of an Autophagy-Related Prognostic Risk Signature for Survival Predicting in Clear Cell Renal Cell Carcinoma Patients. Front Oncol 2020;10:707. [Crossref] [PubMed]
- Sun T. Long noncoding RNAs act as regulators of autophagy in cancer. Pharmacol Res 2018;129:151-5. [Crossref] [PubMed]
- Li D, Li C, Chen Y, et al. LncRNA HOTAIR induces sunitinib resistance in renal cancer by acting as a competing endogenous RNA to regulate autophagy of renal cells. Cancer Cell Int 2020;20:338. [Crossref] [PubMed]
- Su Y, Lu J, Chen X, et al. Long non-coding RNA HOTTIP affects renal cell carcinoma progression by regulating autophagy via the PI3K/Akt/Atg13 signaling pathway. J Cancer Res Clin Oncol 2019;145:573-88. [Crossref] [PubMed]
- Jin Y, Huang R, Xia Y, et al. Long Noncoding RNA KIF9-AS1 Regulates Transforming Growth Factor-β and Autophagy Signaling to Enhance Renal Cell Carcinoma Chemoresistance via microRNA-497-5p. DNA Cell Biol 2020;39:1096-103. [Crossref] [PubMed]
- Lotze MT, Buchser WJ, Liang X. Blocking the interleukin 2 (IL2)-induced systemic autophagic syndrome promotes profound antitumor effects and limits toxicity. Autophagy 2012;8:1264-6. [Crossref] [PubMed]
- Li ML, Xu YZ, Lu WJ, et al. Chloroquine potentiates the anticancer effect of sunitinib on renal cell carcinoma by inhibiting autophagy and inducing apoptosis. Oncol Lett 2018;15:2839-46. [PubMed]
- Messai Y, Noman MZ, Janji B, et al. The autophagy sensor ITPR1 protects renal carcinoma cells from NK-mediated killing. Autophagy 2015; Epub ahead of print. [Crossref] [PubMed]
- Chow PM, Liu SH, Chang YW, et al. The covalent CDK7 inhibitor THZ1 enhances temsirolimus-induced cytotoxicity via autophagy suppression in human renal cell carcinoma. Cancer Lett 2020;471:27-37. [Crossref] [PubMed]