
Discovery of bladder cancer biomarkers in paired pre- and postoperative urine samples
Introduction
Bladder cancer (BC) is a common urological disease, ranking as the seventh most commonly occurring male cancer in urban areas of China in 2015 according to the China Cancer Center (1). The crude and age-standardized by world standard population rates were 5.80/10 and 3.57/10 for incidence (1). Approximately 70% of the newly diagnosed BC cases are non-invasive tumors; however, approximately 50–70% of these patients relapse following surgery, and the probability of the disease developing into invasive BC and endangering life is approximately 30% (2). Urine cytology and cystoscopy are the main tools currently used for the clinical diagnosis of BC, although the sensitivity of urine cytology is poor (3). Cystoscopy has always been considered the gold standard for BC detection, but it is an invasive examination that easily causes patient discomfort (4). The development of novel biomarkers that can aid in comfortable and accurate monitoring for the clinical diagnosis and treatment of BC is urgently needed.
Alterations to protein secretion and expression profiles can occur under different physiological or pathological conditions, with humoral proteomics playing an increasingly important role in the discovery of disease biomarkers. Urine has several unique advantages compared with other body fluids for the development of BC biomarkers. First, urine storage is the main physiological role of the bladder. In BC, the tumor entity is in direct contact with the stored urine and proteins secreted from tumor cells are likely to enter the urine (5,6). Second, proteins in urine are not regulated by homeostasis after urine formation and several potential biomarkers can be enriched (7). Third, urine is often stored in the bladder for some time, and endogenous proteases hydrolyze some of the urinary proteins, so the urine proteome is relatively simple and stable, with low complexity due to the majority of circulating protein being excluded from the kidney tubules during filtration. Fourth, although individual urine samples will, to some extent, have varying protein content and expression profiles, this can be resolved via reasonable methods, such as the use of urinary creatinine normalization and the establishment of a reference urine proteome (8,9).
In previous studies of BC prognosis, biomarker proteins obtained by urinary proteome analysis have been validated against normal controls and general urinary disease control, which has often led to unnecessary biological differences. In the present study, we performed proteomic analysis of paired pre- and postoperative urine samples of BC patients using isobaric tags for relative and absolute quantitation (iTRAQ)-based quantitative technology. The comparison of urine samples from the same patients pre- and postoperatively led to the discovery of significantly differentially expressed proteins, some of which are crucial makers related to the cancer prognosis and may become useful biomarkers for the monitoring and prognosis of BC.
We present the following article in accordance with the MDAR reporting checklist (available at https://dx.doi.org/10.21037/tau-21-562).
Methods
Study population and sample collection
All urine samples were obtained from the pathology biosample bank of the Fifth Medical Centre of Chinese PLA General Hospital. The urine samples were collected between January 2018 and July 2018 and stored at −80 °C prior to analysis. Overall, 40 urine samples were used in this study: 8 samples from volunteers with no history or evidence of BC; 4 from patients with general urinary system diseases; 16 from patients with BC prior to surgery; and 12 samples from patients with BC following surgical removal of their tumor. We confirmed that participants did not present symptoms of kidney failure (for the details of pathology information, see https://cdn.amegroups.cn/static/public/tau-21-562-1.xlsx). The presence of BC was confirmed using cystoscopy, together with histopathological information obtained following subsequent surgical interventions. Tumors were graded according to American Joint Committee on Cancer (AJCC) tumor, node, metastasis (TNM) criteria and clinical stages. An immunohistochemical (IHC) tissue chip, containing fixed samples of normal and cancerous bladder tissue, was purchased from US Biomax (BL481c, Rockville, MD, USA). All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the ethics committee of the Fifth Medical Centre of Chinese PLA General Hospital (NO.ky-2018-2-13), and informed consent was taken from all the patients.
Experimental design
The overall experimental workflow is shown in Figure 1. Urine samples were collected pre- and postoperatively and were immediately stored at −80 °C. Samples were thawed at 4 °C, followed by centrifugation at 3,000 rpm to remove obvious precipitation. The supernatant was concentrated by ultrafiltration using a 3-kD ultrafiltration tube, quantified by bicinchoninic (BCA) assay, and stored at −80 °C. Proteins within the sample were digested using trypsin (1:50) at 37 °C overnight. The digested peptides were desalted using hydrophilic-lipophilic balanced (HLB) cartridges, and then labeled with the iTRAQ reagent kit following the manufacturer’s protocol (8plex, Applied Biosystems, Bedford, MA, USA). iTRAQ is a powerful technique of protein quantification, which can utilize a multiplexed isobaric chemical tagging reagent for identification and quantitation of proteins in multiple samples at the same time. The pre-operative urine samples were labeled with 113, 115, 117, and 119, and the postoperative urine samples were labeled with 114, 116, 118, and 121, respectively. Two iTRAQ experiments were implemented, and the urine samples from different patients represent the different biological replicates. Subsequently, an equal ratio of each set was pooled and analyzed by a Q-Exactive mass spectrometry (Thermo Fisher Scientific, Waltham, MA, USA). Candidate biomarkers from proteomic analysis were simultaneously validated using western blotting (WB) in independent cohorts and IHC staining, and combined with gene expression data from BC samples in TCGA to screen out the final biomarkers.
Data processing
The tandem mass spectrometry (MS/MS) data were processed using MaxQuant software (version 1.6.0.13, https://www.maxquant.org) and searched against the SwissProt database (Homo sapiens, 2017_09 version) (10,11). Specific cleavages were selected for trypsin, and the precursor and fragment ion mass tolerance were set to 20 ppm and 0.02 Da, respectively. The carbamidomethyl of cysteine, iTRAQ-8plex of lysine, and N-terminal were stated as constant modifications, while the oxidation of methionine, acetyl of the N-terminal constituted the variable modifications. The default target-decoy approach with the reverse sequence database was used and a false discovery rate (FDR) <0.01 at both peptide and protein level were applied. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the iProX (12) partner repository with the dataset identifier PXD011871.
Statistical and bioinformatics analysis
The quality-controlled data first used the Shapiro-Wilk test to verify the assumption of normality. If the assumption was met, the data is compared using paired t-test. If the normality assumption was not met, the data was subjected to Wilcoxon signed-rank test. Fold change (FC) was obtained by comparing preoperative with postoperative protein expression. The differentially expressed urinary proteins were defined as those with a P value <0.05 and an FC >1.50 or <0.67 in groups between pre- and postoperative urine samples. If more than 75% of the samples met the above criteria, then the relevant protein was be listed as a candidate marker list.
Ingenuity pathway analysis (IPA) was used to classify differentially expressed proteins (DEPs) according to functional annotations (13), including cellular components, metabolic processes, and classical pathways. Functional annotation and pathway analysis of DEPs could help screen for appropriate candidate biomarkers with relevance to BC cellular processes.
WB
We performed WB of urine samples to validate the MS results. First, we used Coomassie brilliant blue staining to quantify the total protein content of each urine sample, which was used as an internal reference for loading the sample for WB (14). The samples were separated sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to polyvinylidene fluoride (PVDF) membranes. After blocking, the membranes were incubated with primary antibodies against cadherin-related family member 2 (CDHR2, Proteintech, 27103-1-AP), heat shock protein (HSP) beta-1 (HSP27, Proteintech, 18284-1-AP), or alpha-1-antitrypsin (A1AT, Proteintech, 16382-1-AP) at 4 °C overnight. Following washing, the membranes were incubated with horseradish peroxidase (HRP)-conjugated mouse anti-rabbit-IgG, heavy-chain-specific secondary antibody or HRP-conjugated mouse anti-rabbit-IgG, light-chain-specific secondary antibody to avoid non-specific binding to potential immunoglobulins due to the patient’s hematuria. Finally, blots were developed using an enhanced chemiluminescence system. ImageJ analysis software (National Institutes of Health, Bethesda, MD, USA) was used to analyze the intensity of each band.
Immunohistochemistry staining
The BC tissue chip (BL481c) was dewaxed and hydrated, then boiled in 10 M citrate buffer solution for antigen retrieval; endogenous peroxidases were blocked using 3% hydrogen peroxide. The slides were incubated with primary antibodies [CDHR2 (27103-1-AP, 1:200), HSP27 (18284-1-AP, 1:450), SFN (66251-1-Ig, 1:200), and LMNA (YC0057, 1:200)] in a humid box at 4 °C overnight. Negative controls were generated by omitting the primary antibody. After washing, the slides were incubated with HRP-conjugated goat anti-rabbit secondary antibody. After the incubation, the colorimetric reaction was performed using 3,3-diaminobenzidine reagent, and nuclei were counterstained using hematoxylin. Slides were washed with water for 40 min, dehydrated after bluing, and sealed with a neutral gum. The prepared slides were analyzed by a qualified pathologist.
Results
Urine proteomics pre- and postoperatively in BC patients
To determine the reliability of urinary biomarkers, we performed two independent mass spectrometry experiments—the first from urine samples collected within one week postoperatively, and the second from urine samples collected over two weeks postoperatively. Overall, 1,162 proteins were co-identified in urine samples obtained at two time points (Figure 2A), and the complete list of proteins was tabulated (https://cdn.amegroups.cn/static/public/tau-21-562-2.xlsx). Simultaneously, we also found some tumor markers mentioned in the previous literature, such as alpha-1 antitrypsin (A1AT), Lithostathine-1-alpha (REG1A), gastrotropin (FABP6), and apolipoprotein C-I (APOC1) (15-18). Within one week postoperatively, there were 75 DEPs. Over two weeks postoperatively, there were 184 DEPs.
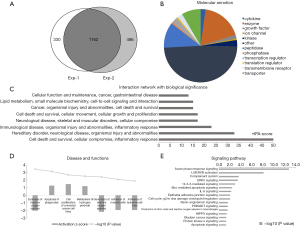
Interestingly, despite that numerous proteins were co-identified at both postoperative time points; we found that there was little overlap between the DEPs at the two time points. This could be attributed to the postoperative time during which the urine was collected, with the acute phase stress response of the patients exerting a substantial effect on the postoperative protein profile. We also confirmed this inference in the mass spectrometry data, which revealed that acute phase response signaling proteins were significantly enriched in the DEPs at both time points, and confirmed that the urine sample proteomic profile does indeed reflect changes in the body following tumor resection. After considering the postoperative acute phase response, we believe that the DEPs over two weeks postoperatively provide a more accurate list of tumor biomarker candidate proteins than those within one week postoperatively.
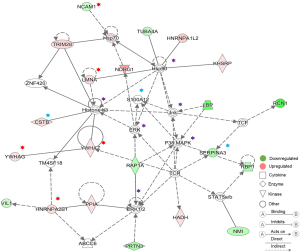
In order to gain a deeper understanding of the DEPs at two weeks later between pre- and postoperative urine samples, we used IPA to perform bioinformatics analysis of DEPs, and the results also demonstrated the reliability of our screening results. At two weeks after operation, we saw that a large number of DEPs were classified as cytokines, enzymes, transcriptional regulators, transporters, and so on (Figure 2B). The interaction network of DEPs was mainly concentrated in cell death and survival, inflammatory response, cellular growth and proliferation, lipid metabolism, and so on (Figure 2C). The biological functions and classical pathways involved were related to the survival and development of cancer (Figure 2D,E). For example, the pathways that related to BC signaling and tumor development were identified, including HIPPO signaling, 14-3-3-mediated signaling, and cell cycle (G2/M DNA damage checkpoint) regulation. Molecular interaction network analysis also showed that the screened DEPs were closely related to known cancer marker proteins (Figure 3).
Differential protein expression pre- and postoperatively in BC patients
For the 184 DEPs obtained over two weeks postoperatively, we filtered out all immunoglobulin (IgG) proteins to avoid the effects of hematuria in patients with BC. We used the UniProt database annotation to perform a preliminary screening of the remaining 101 proteins differentially expressed between pre- and postoperative BC samples in terms of the functions of the protein and its correlation with tumorigenesis, together with known related literature. We obtained a list of candidate biomarker proteins for BC (for the complete list of DEPs, see https://cdn.amegroups.cn/static/public/tau-21-562-3.xlsx). Below, we highlight the potential role of some selected DEPs in BC: cadherin-related family member 2 (CDHR2), HSP beta-1 (HSP27), prelamin-A/C (LMNA), 14-3-3 protein sigma (SFN), 14-3-3 protein zeta/delta (YWHAZ), 14-3-3 protein gamma (YWHAG), and heterogeneous nuclear ribonucleoproteins A2/B1 (HNRNPA2B1).
The intercellular adhesion factor and member of the cadherin family, CDHR2, showed low expression prior to surgery. Consistent with our MS result, previous studies have shown that CDHR2 expression is significantly reduced in cancer from liver, kidney, and colon compared to its expression in these tissues under normal conditions (19). It may be that CDHR2 inhibits tumor formation via interaction with hMAST205 (microtubule-associated serine/threonine protease-205KDa) to induce contact inhibition of epithelial cells (20), or via interaction with β-catenin to inhibit signal transduction and inhibit tumor cell proliferation (21).
Also known as HSPB1, HSP27 is highly expressed in the urine of patients with BC prior to surgery. Consistent with our MS result, heat shock protein (HSP) is often expressed at high levels in tumors that can rapidly respond to cellular stress. The HSP promotes cancer cell proliferation, reduces cancer cell growth inhibition, and promotes tumor angiogenesis (22). The HSP27 can significantly enhance smooth muscle cell proliferation as well as can contribute to the tumor cell infiltration process (23). Large-scale clinical samples have confirmed that high HSP27 expression is associated with BC prognosis and progression (24).
A precursor of the nuclear membrane component lamin, LMNA shows high expression preoperatively. The imbalance of lamin expression may be directly related to the rapid mitotic division of cancer cells. Studies have shown that the differential LMNA expression in prostate cancer tissues with low and high Gleason scores may be closely related to epithelial-mesenchymal (EMT) transformation (25). In colon cancer, the fusion of LMNA with high-affinity nerve growth factor receptor NTRK1 has been found to induce metastasis (26).
All having high preoperative expression, SFN, YWHAG, and YWHAZ are members of the 14-3-3 protein family, a class of important protein signaling pathway regulators that are ubiquitous and highly conserved in eukaryotes. The SFN can regulate protein synthesis and epithelial cell growth when binding to the keratin protein KRT17 (27), and SFN overexpression prevents cell migration and enhances mitogen-activated protein kinase (MAPK) signaling in wound healing (28). However, the reported results from literatures are inconsistent with our mass spectrometry results showing high SFN expression in preoperative urine samples. It may be that SFN binds to BCL2L1 to inhibit cell death, which may explain the phenomenon of high SFN expression in cancer (Figure S1).
The high preoperative YWHAG and YWHAZ expression was consistent with previous studies showing that YWHAG overexpression induces activation of the phosphatidylinositol 3-kinase (PI3K) and MAPK pathways (29). Moreover, YWHAG is highly expressed in hepatocellular carcinoma and breast cancer and has been suggested as a target for cancer treatment (30,31). Amplification of YWHAZ in the genome is also common in patients with BC in the TCGA database (32). It has also been suggested that knocking down YWHAZ promotes tumorigenesis in vitro and in vivo in BC (33). It is possible that YWHAZ plays an anti-tumor role in the appearance of BC, but other molecular mechanisms may be involved in tumor development.
In addition, we identified a series of low-abundance proteins, such as DNA damage-binding protein 1, which may be involved in DNA repair during tumor cell development (34). We found the expressions of 60S ribosomal protein L12, adenylate kinase isoenzyme 1, heterogeneous nuclear ribonucleoproteins A2/B1, and lactoylglutathione lyase were higher in postoperative samples than those in preoperative samples, that may be involved in the protein and energy synthesis functions of tumor cells (35,36). Additionally, we found high preoperative expressions of nuclease-sensitive element-binding protein 1 and eukaryotic translation initiation factor 5A, which could improve the transcription efficiency, promote cell division, and stimulate cell migration (37). High preoperative expression of ectonucleotide pyrophosphatase/phosphodiesterase family member 2 may stimulate tumor movement and promote angiogenesis (38). Low preoperative expression of neural cell adhesion molecule 1 may facilitate tumor cell migration (39).
Validation of differential protein expression using WB and IHC
Considering the symptoms of hematuria in patients with BC, enzyme-linked immunosorbent assay (ELISA) may lead to partial false positive results. We used WB for verification experiments. Given the abundance changes of candidate proteins pre- and postoperatively, and technical difficulty, we selected 4 proteins (HSP27, CDHR2, SFN, LMNA) for biomarker validation via WB and IHC (for the complete information of IHC, see https://cdn.amegroups.cn/static/public/tau-21-562-4.xlsx). The results of WB are discussed below (Figure 4).
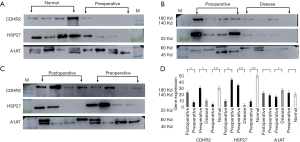
To validate the mass spectrometry results by WB, we established the following verification comparisons: preoperative group vs. healthy control group (Figure 4A), preoperative group vs. postoperative group (Figure 4B), and preoperative group vs. general urinary system disease group (Figure 4C). Before the experiment, we used Coomassie brilliant blue staining to quantify the total protein in urine samples which we then used as an internal reference for WB (Figure S2). The WB membranes were incubated with an HRP-conjugated mouse anti-rabbit-IgG heavy-chain-specific or light-chain-specific secondary antibody to avoid non-specific binding to potential immunoglobulins due to the patient’s hematuria (Figure S3). Validation with western blotting was followed by validation via immunostaining on a fixed bladder tissue sample chip containing normal, non-invasive cancer, and invasive cancer bladder tissue.
In WB, CDHR2 expression was significantly different in the preoperative group compared with the postoperative group and in the preoperative group compared with the normal control group (Figure 4D). In subsequent IHC validation (Figure 5A), CDHR2 expression was significantly decreased in 14 cases of low-grade urothelial carcinomas compared with normal bladder tissues (P=0.01), and CDHR2 expression level did not significantly differ in 24 cases of high-grade urothelial carcinomas (P=0.071). No significant change was observed in CDHR2 expression in 38 cases of BC tissues compared with normal bladder tissues (P=0.079). Although CDHR2 expression level showed no significant differences between invasive BC tissues and normal bladder tissue, a significant decrease in CDHR2 expression levels often predicted the presence of invasive BC.
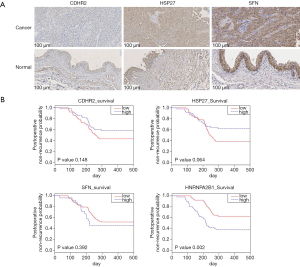
The WB showed high HSP27 expression in the normal control samples, which was unexpected given the high HSP27 expression in preoperative urine samples displayed in the mass spectrometry results (Figure 4D). The preoperative urine samples tended to show higher expression compared with postoperative urine samples, but the preoperative urine samples showed significantly lower HSP27 expression compared with the normal control samples. In the subsequent IHC validation (Figure 5A), HSP27 expression was not definitively associated with cancer; among the 40 cases of BC, 13 (32.5%) showed high HSP27 expression, 21 (52.5%) did not significantly differ from normal bladder tissue, and 6 samples (15%) tended to have lower HSP27 expression than normal bladder tissue. A significant increase in HSP27 expression levels often predicted the presence of invasive BC.
In the WB experiment, SFN and LMNA were failed to obtain significant bands. In the subsequent IHC validation, a total of 46 bladder samples were involved. The tissue microarray of SFN included 24 high-grade urothelial carcinomas, 14 low-grade urothelial carcinomas, and 8 normal bladder tissues. The results of IHC showed that SFN was highly expressed in normal tissues (P=0.018), which was consistent with previous literature records (Figure 5A). The LMNA tissue chip included 22 high-grade urothelial carcinomas, 16 low-grade urothelial carcinomas, and 8 normal bladder tissues. No statistically significant results were obtained in the LMNA results, but the comparison of high-grade urothelial carcinoma with normal tissues showed that LMNA has a high expression trend in high-grade urothelial carcinoma (Figure S4A).
Validation of differential protein expression using TCGA BC gene expression data
We incorporated BC gene expression data from TCGA to verify the relationship between protein expression and BC recurrence time (Figure 5B, Figure S4B). The relationship between the selected oncogene expression and the postoperative recurrence time of BC supported the results of our mass spectrometry and WB experiments (https://cdn.amegroups.cn/static/public/tau-21-562-5.xlsx). The candidate biomarkers could predict the prognosis of patients 200 days postoperatively. Although only HNRNPA2B1 expression was significantly associated with the recurrence rate of BC, except HSP27, the relationship between the expression trends of other biomarkers and the postoperative recurrence rate were consistent with our expectation of mass spectrometry and WB data.
Discussion
As a form of urological cancer with a high recurrence rate, a large proportion of BC patients develop invasive BC. Effective monitoring of cancer recurrence in patients with BC is of the utmost importance following initial surgical treatment (40). Currently, reliable biomarkers of urinary system diseases are effective supplements or alternative tests for cytology and cystoscopy (15,41). In the present study, we determined the pre- and postoperative proteomic data of urine samples obtained from the same patients. The data from the 2 postoperative time points confirmed the occurrence of the above-mentioned postoperative stress response. For example, protein angiotensinogen (AGT) as a negative regulator of angiogenesis (42), and fibrinogen beta and gamma chains, previously identified as a tumor biomarker (43,44) but which can also play a role in blood vessel contraction and hemostasis, were both highly expressed postoperatively. The pre- and postoperative levels of A1AT and APOC1 appeared to be inconsistent with their known function as BC biomarkers, but their expression was consistent at both postoperative time points. This may be because of the postoperative requirement for several cell proliferation and angiogenesis factors to aid the body heal the wound and resist the invasion of pathogens (45,46). Moreover, most of the proteins such as A1AT and APOC1 can be derived from blood, which may be present in postoperative urine samples.
To further confirm that the specificity of the postoperative urine samples is the cause of this phenomenon, we used WB experiments to confirm A1AT expression, which is known to be highly associated with cancer (Figure 4). The abundance of A1AT we measured in both WB and mass spectrometry experiments was not consistent with previous literature (47,48). The IHC results of SFN also reflected similar problems. Compared with BC tissues, SFN showed a high expression trend in normal tissues, but the abundance of SFN in postoperative urine was lower than that in preoperative urine. Therefore, we believe that the control sample and postoperative sample have different protein profiles, both in tissues and urine. Thus, the postoperative sample could be a better resource for discovery of biomarkers.
By comparison of the urine samples from the same patients pre- and postoperatively, we identified a list of candidate biomarker proteins for BC, and some of them may play important roles in tumorigenesis and development. For example, CDHR2, showed low expression in preoperative urine of BC patients, which may be effective in inhibiting signal transduction and tumor cell proliferation in BC. HSP27 were highly expressed in the urine of BC patients prior to surgery, which can significantly enhance smooth muscle cell proliferation as well as contribute to the tumor cell infiltration process.
In summary, we employed urine proteomics to successfully identify BC prognostic biomarkers. Moreover, we completed the initial validation of these markers in BC samples using molecular experimental techniques or gene expression data from TCGA. We believe that following extensive cohort validation, some of these proteins could be helpful in screening for BC prognosis.
Acknowledgments
The authors are grateful for the help provided by the Bioinformatics Laboratory in the National Center for Protein Sciences (Beijing). We thank the Pathology Department of the Fifth Medical Centre of Chinese PLA General Hospital for providing pathological analysis. The authors would also like to express their gratitude to all volunteers who provided samples in this experiment.
Funding: This work was supported by the National Key Research and Development Program of China (2020YFE0202200).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://dx.doi.org/10.21037/tau-21-562
Data Sharing Statement: Available at https://dx.doi.org/10.21037/tau-21-562
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/tau-21-562). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the ethics committee of the Fifth Medical Centre of Chinese PLA General Hospital (No. ky-2018-2-13), and informed consent was taken from all the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Zhang S, Sun K, Zheng R, et al. Cancer incidence and mortality in China, 2015. J Nat Cancer Cent 2021;1:2-11. [Crossref]
- Zang Y, Li X, Cheng Y, et al. An overview of patients with urothelial bladder cancer over the past two decades: a Surveillance, Epidemiology, and End Results (SEER) study. Ann Transl Med 2020;8:1587. [Crossref] [PubMed]
- Urquidi V, Rosser CJ, Goodison S. Molecular diagnostic trends in urological cancer: biomarkers for non-invasive diagnosis. Curr Med Chem 2012;19:3653-63. [Crossref] [PubMed]
- Black AJ, Black PC. Variant histology in bladder cancer: diagnostic and clinical implications. Transl Cancer Res 2020;9:6565-75. [Crossref]
- Frantzi M, Latosinska A, Flühe L, et al. Developing proteomic biomarkers for bladder cancer: towards clinical application. Nat Rev Urol 2015;12:317-30. [Crossref] [PubMed]
- Hentschel AE, van der Toom EE, Vis AN, et al. A systematic review on mutation markers for bladder cancer diagnosis in urine. BJU Int 2021;127:12-27. [Crossref] [PubMed]
- Zhu CZ, Ting HN, Ng KH, et al. A review on the accuracy of bladder cancer detection methods. J Cancer 2019;10:4038-44. [Crossref] [PubMed]
- Leng W, Ni X, Sun C, et al. Proof-of-Concept Workflow for Establishing Reference Intervals of Human Urine Proteome for Monitoring Physiological and Pathological Changes. EBioMedicine 2017;18:300-10. [Crossref] [PubMed]
- Stanley S, Vanarsa K, Soliman S, et al. Comprehensive aptamer-based screening identifies a spectrum of urinary biomarkers of lupus nephritis across ethnicities. Nat Commun 2020;11:2197. [Crossref] [PubMed]
- Tyanova S, Temu T, Cox J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat Protoc 2016;11:2301-19. [Crossref] [PubMed]
- UniProt Consortium T. UniProt. the universal protein knowledgebase. Nucleic Acids Res 2018;46:2699. [Crossref] [PubMed]
- Ma J, Chen T, Wu S, et al. iProX: an integrated proteome resource. Nucleic Acids Res 2019;47:D1211-7. [Crossref] [PubMed]
- Krämer A, Green J, Pollard J Jr, et al. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 2014;30:523-30. [Crossref] [PubMed]
- Schreiber J, Végh MJ, Dawitz J, et al. Ubiquitin ligase TRIM3 controls hippocampal plasticity and learning by regulating synaptic γ-actin levels. J Cell Biol 2015;211:569-86. [Crossref] [PubMed]
- Xylinas E, Kluth LA, Rieken M, et al. Urine markers for detection and surveillance of bladder cancer. Urol Oncol 2014;32:222-9. [Crossref] [PubMed]
- Li H, Li C, Wu H, et al. Identification of Apo-A1 as a biomarker for early diagnosis of bladder transitional cell carcinoma. Proteome Sci 2011;9:21. [Crossref] [PubMed]
- Lin SY, Chang CH, Wu HC, et al. Proteome Profiling of Urinary Exosomes Identifies Alpha 1-Antitrypsin and H2B1K as Diagnostic and Prognostic Biomarkers for Urothelial Carcinoma. Sci Rep 2016;6:34446. [Crossref] [PubMed]
- Ohlsson G, Moreira JM, Gromov P, et al. Loss of expression of the adipocyte-type fatty acid-binding protein (A-FABP) is associated with progression of human urothelial carcinomas. Mol Cell Proteomics 2005;4:570-81. [Crossref] [PubMed]
- Ose R, Oharaa O, Nagase T. Galectin-1 and Galectin-3 Mediate Protocadherin-24-Dependent Membrane Localization of β-catenin in Colon Cancer Cell Line HCT116. Curr Chem Genomics 2012;6:18-26. [Crossref] [PubMed]
- Okazaki N, Takahashi N, Kojima S, et al. Protocadherin LKC, a new candidate for a tumor suppressor of colon and liver cancers, its association with contact inhibition of cell proliferation. Carcinogenesis 2002;23:1139-48. [Crossref] [PubMed]
- Ose R, Yanagawa T, Ikeda S, et al. PCDH24-induced contact inhibition involves downregulation of beta-catenin signaling. Mol Oncol 2009;3:54-66. [Crossref] [PubMed]
- O'Callaghan-Sunol C, Gabai VL, Sherman MY. Hsp27 modulates p53 signaling and suppresses cellular senescence. Cancer Res 2007;67:11779-88. [Crossref] [PubMed]
- Wang Y, Xiong Z, Gong W, et al. Expression of heat shock protein 27 correlates with actin cytoskeletal dynamics and contractility of cultured human bladder smooth muscle cells. Exp Cell Res 2015;338:39-44. [Crossref] [PubMed]
- Yu HJ, Chang YH, Pan CC. Prognostic significance of heat shock proteins in urothelial carcinoma of the urinary bladder. Histopathology 2013;62:788-98. [Crossref] [PubMed]
- Zuo L, Zhao H, Yang R, et al. Lamin A/C might be involved in the EMT signalling pathway. Gene 2018;663:51-64. [Crossref] [PubMed]
- Sartore-Bianchi A, Ardini E, Bosotti R, et al. Sensitivity to Entrectinib Associated With a Novel LMNA-NTRK1 Gene Fusion in Metastatic Colorectal Cancer. J Natl Cancer Inst 2016;108:djv306 [Crossref] [PubMed]
- Yang W, Dicker DT, Chen J, et al. CARPs enhance p53 turnover by degrading 14-3-3sigma and stabilizing MDM2. Cell Cycle 2008;7:670-82. [Crossref] [PubMed]
- Benzinger A, Muster N, Koch HB, et al. Targeted proteomic analysis of 14-3-3 sigma, a p53 effector commonly silenced in cancer. Mol Cell Proteomics 2005;4:785-95. [Crossref] [PubMed]
- Radhakrishnan VM, Putnam CW, Martinez JD. Activation of phosphatidylinositol 3-kinase (PI3K) and mitogen-activated protein kinase (MAPK) signaling and the consequent induction of transformation by overexpressed 14-3-3γ protein require specific amino acids within 14-3-3γ N-terminal variable region II. J Biol Chem 2012;287:43300-11. [Crossref] [PubMed]
- Choi JE, Hur W, Jung CK, et al. Silencing of 14-3-3ζ over-expression in hepatocellular carcinoma inhibits tumor growth and enhances chemosensitivity to cis-diammined dichloridoplatium. Cancer Lett 2011;303:99-107. [Crossref] [PubMed]
- Neal CL, Yao J, Yang W, et al. 14-3-3zeta overexpression defines high risk for breast cancer recurrence and promotes cancer cell survival. Cancer Res 2009;69:3425-32. [Crossref] [PubMed]
- Liu S, Wu Y, Yang T, et al. Coexistence of YWHAZ amplification predicts better prognosis in muscle-invasive bladder cancer with CDKN2A or TP53 loss. Oncotarget 2016;7:34752-8. [Crossref] [PubMed]
- Liu S, Jiang H, Wen H, et al. Knockdown of tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein zeta (YWHAZ) enhances tumorigenesis both in vivo and in vitro in bladder cancer. Oncol Rep 2018;39:2127-35. [Crossref] [PubMed]
- Jin J, Arias EE, Chen J, et al. A family of diverse Cul4-Ddb1-interacting proteins includes Cdt2, which is required for S phase destruction of the replication factor Cdt1. Mol Cell 2006;23:709-21. [Crossref] [PubMed]
- Imami K, Milek M, Bogdanow B, et al. Phosphorylation of the Ribosomal Protein RPL12/uL11 Affects Translation during Mitosis. Mol Cell 2018;72:84-98.e9. [Crossref] [PubMed]
- Hu Y, Sun Z, Deng J, et al. Splicing factor hnRNPA2B1 contributes to tumorigenic potential of breast cancer cells through STAT3 and ERK1/2 signaling pathway. Tumour Biol 2017;39:1010428317694318 [Crossref] [PubMed]
- Mathews MB, Hershey JW. The translation factor eIF5A and human cancer. Biochim Biophys Acta 2015;1849:836-44. [Crossref] [PubMed]
- Quan M, Cui JJ, Feng X, et al. The critical role and potential target of the autotaxin/lysophosphatidate axis in pancreatic cancer. Tumour Biol 2017;39:1010428317694544 [Crossref] [PubMed]
- Deborde S, Omelchenko T, Lyubchik A, et al. Schwann cells induce cancer cell dispersion and invasion. J Clin Invest 2016;126:1538-54. [Crossref] [PubMed]
- Peak TC, Hemal A. Partial cystectomy for muscle-invasive bladder cancer: a review of the literature. Transl Androl Urol 2020;9:2938-45. [Crossref] [PubMed]
- Chou R, Gore JL, Buckley D, et al. Urinary Biomarkers for Diagnosis of Bladder Cancer: A Systematic Review and Meta-analysis. Ann Intern Med 2015;163:922-31. [Crossref] [PubMed]
- Lin J, Chen J, Liu C. AGT M235T variant is not associated with risk of cancer. J Renin Angiotensin Aldosterone Syst 2015;16:448-52. [Crossref] [PubMed]
- Lindén M, Segersten U, Runeson M, et al. Tumour expression of bladder cancer-associated urinary proteins. BJU Int 2013;112:407-15. [Crossref] [PubMed]
- Duan S, Gong B, Wang P, et al. Novel prognostic biomarkers of gastric cancer based on gene expression microarray: COL12A1, GSTA3, FGA and FGG. Mol Med Rep 2018;18:3727-36. [Crossref] [PubMed]
- Raza A, Franklin MJ, Dudek AZ. Pericytes and vessel maturation during tumor angiogenesis and metastasis. Am J Hematol 2010;85:593-8. [Crossref] [PubMed]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011;144:646-74. [Crossref] [PubMed]
- Miyake M, Ross S, Lawton A, et al. Investigation of CCL18 and A1AT as potential urinary biomarkers for bladder cancer detection. BMC Urol 2013;13:42. [Crossref] [PubMed]
- Shakya R, Tarulli GA, Sheng L, et al. Mutant p53 upregulates alpha-1 antitrypsin expression and promotes invasion in lung cancer. Oncogene 2017;36:4469-80. [Crossref] [PubMed]
(English Language Editor: J. Jones)


