
CYR61, regulated by miR-22-3p and MALAT1, promotes autophagy in HK-2 cell inflammatory model
Introduction
Acute kidney injury (AKI) is a common clinical critical disease with high incidence, high mortality, and low diagnosis rate (1). The process of tubular injury and repair plays a key role in epithelial-mesenchymal transition (EMT), renal fibrosis, acute renal injury-chronic transition, and the progression of nephropathy (2-4). The degree of renal tubular injury is closely related to the degree of renal function decline (5). Due to the strong reabsorption function of renal tubules, tubular epithelial cells have high metabolic activity. energy demand, and can secrete a variety of cytokines. In various diseases, renal tubular epithelial cells are prone to structural and functional abnormalities. On the one hand, because they are in direct contact with urine, the proteins and cytokines in urine directly cause cellular damage or phenotypic transformation and release various inflammatory factors and growth factors. On the other hand, tubular epithelial cells are closely structurally connected to the renal interstitium, and damaged cells can be directly involved in interstitial inflammation and fibrosis, or play an important role in the process of fibrosis by recruiting inflammatory cells and promoting the proliferation of intrinsic cells in the interstitium. Therefore, protecting the structural and functional integrity of renal tubular epithelial cells in the inflammatory state is an effective therapy to slow the progression of nephropathy.
As the component of gram-negative bacteria cell walls, lipopolysaccharide (LPS) is a common endotoxin and is often used as an inducer for inflammatory modeling in vitro and in vivo. It has been reported that LPS can induce acute nephritis in mice, and can also induce the secretion of inflammatory factors such as interleukin (IL)-1β, interleukin 6 (IL-6), and tumor necrosis factor-α (TNF-α) in cultured human renal tubular epithelial cells (HK-2) in vitro (6).
Recent studies have reported that Cysteine Rich Protein 61 (CYR61, also known as cellular communication network factor 1, CCN1) could regulate autophagy and apoptosis both in vivo and in vitro (7,8). By binding to integrin ligand and participating in a variety of signaling pathways, CYR61 plays a variety of physiological functions (9,10). Purified CYR61 has been shown to promote proliferation, migration, and adhesion of endothelial cells, enhanced fibroblast growth factor and platelet-derived growth factor-induced DNA synthesis in fibroblasts, stimulated chondrocyte growth and differentiation, and promote angiogenesis (11,12). Additionally CYR61 has been shown to inhibit fibrosis in the repair of liver, lung, skin, heart, and other tissues (13-15). However, in a model of acute obstructive renal fibrosis, CYR61 participated in the inflammatory response induced by transforming growth factor-p (TGF-p), and inhibition of CYR61 expression improved interstitial inflammation (16). After ischemia, CYR61 was increased in human renal tissue, and was involved in angiogenesis, positive regulation of movement, and monomer cell adhesion (17). Therefore, the role of CYR61 in renal tubular epithelial cells in an inflammatory state and its upstream regulatory factors requires further exploration. It has been shown that miR-22-3p inhibits LPS-induced acute renal injury in vivo (18).The inhibition of long non-coding RNA (lncRNA) MALAT1, which is involved in various pathological processes such as tumor growth and inflammation (19), impaired the malignant behavior of AKI (20,21). Through sponging miR-22-3p, MALAT1 exerts a negative regulatory effect (22).
Both miRNA-22-3p and MALAT1 regulate the secretion of inflammatory factors, apoptosis, and autophagy in a nephritis cell model. We concluded that if CYR61 is involved in autophagy and apoptosis of renal tubular dermal cells in an inflammatory state, its upstream regulatory factors could be miRNA-22-3p and MALAT1. To verify this hypothesis, we first constructed an inflammation model of HK-2 cells induced by LPS and verified whether CYR61 was involved in autophagy and apoptosis under inflammatory state by short hairpin RNA (shRNA). Subsequently, the regulation relationship between miRNA-22-3p and CYR61 was detected by dual-luciferase assay, and the regulation of miRNA-22-3p on CYR61 and the mutual regulation relationship between miRNA-22-3p and MALAT1 were further verified by overexpression vector. Through the above experiments, we hoped to clarify the role of CYR61 in the process of autophagy and apoptosis of renal tubular dermal cells in a state of inflammation and explore its upstream regulatory molecular mechanism.
We present the following article in accordance with the MDAR reporting checklist (available at https://dx.doi.org/10.21037/tau-21-623).
Methods
Cell culture and LPS-induced inflammation model
The HK-2 and HEK293T cell lines were obtained from Chinese Tissue Culture Collections (CTCC cat. No. CTCC-CL-375, CTCC-CL-379, Jinhua, Zhejiang, China), Cells were cultured in Roswell Park Memorial Institute (RPMI) 1640 medium (cat. No. A1049101, Gibco, Inc., Amarillo, TX, USA) with 10% fetal bovine serum (FBS; cat. No. 16140071, Gibco, Inc. USA), and 1% streptomycin/penicillin (cat. No. E607011, Sangon, Inc., Shanghai, China), and were maintained at 37 °C in an atmosphere of 5% CO2 in air.
In order to explore the appropriate conditions of the LPS-induced HK-2 cell inflammation model, the HK-2 cell concentration was adjusted to 5×104 cells/mL, then the cells were inoculated into 96-well plates, 100 µL per well. After cultured for 24 h, cells were treated with different concentrations of LPS (0, 0.01, 0.1, 1, 10 µg/mL, cat. No. P1400, Solarbio, Inc., Beijing, China), and cultured for 8, 12, and 24 h, respectively. Then Cell Counting Kit-8 (CCK-8) assays were performed. The results of pre-experiments (Figure S1) showed that HK-2 cell activity decreased significantly after 12 h treatment with 1 µg/mL LPS, so this condition was selected for subsequent experiments.
Transfection
Lentivirus (Oligobio, Inc., China) was used to transfect shRNA. In order to find the best multiplicity of infection (MOI) for transfection, lentiviruses with different MOI (10, 50, 100) were used to infect the cells. After 48 h culture, the efficiency of virus infection was observed under a fluorescence microscope. The results of pre-experiments (Figure S2) showed that the infection efficiency reached more than 95% with an MOI value of 50. Therefore, the MOI value of 50 was selected for the subsequent experiment.
To transfect miRNAs or vectors, Lipofectamine 2000 (cat. No. 11668027, Thermo Fisher, Inc., Waltham, MA, USA) was mixed with Dulbecco’s modified eagle medium (DMEM)-H (cat. No. 11965092, Gibco, Inc., USA) containing miRNAs or plasmids to 25 µL, incubated at room temperature for 20 min, and then added to the cell medium. After 6 h of culture, the medium was replaced with a complete medium and cultured for 48 h before subsequent experiments.
Electron microscopic analysis
The cells were fixed with 2.5% glutaraldehyde and washed with phosphate-buffered saline (PBS) 3 times, then fixed with 1% osmium acid at room temperature (20 °C) for 2 h. After washing, the cells were dehydrated with alcohol. We used 812 embedding agent (cat. No. 45359, Merck, Inc., Kenilworth, NJ, USA) for permeation and embedding. Then, the samples were polymerized at 60 °C for 48 h and sliced into sections at 60–80 nm. The sections were double-stained with lead and uranium before being observed using a transmission electron microscope (HT7700-SS, Hitachi, Tokyo, Japan).
Quantitative reverse transcription polymerase chain reaction
Total RNA was extracted using Trizol reagent. then reverse transcribed into DNA using a reverse transcription polymerase chain reaction (RT-PCR) kit (cat. No. K1622, Thermo Fisher, Inc. USA), according to the manufacturer’s instructions. Real-time PCR (qPCR) was performed with a SYBR Green PCR kit (cat. No. F-415XL, Thermo, Inc. USA) on a qPCR system (ABI-7500, Applied Biosystems, Waltham, MA, USA) with glyceraldehyde 3-phosphate dehydrogenase (GAPDH) used as internal control (23). The 2−∆∆Ct method was used to analyze the data (24). The primers for qPCR were purchased from Sangon Biotech Company. The sequences of the primers are shown in Table S1.
Western blot (WB) assay
Cellular proteins were extracted using radioimmunoprecipitation assay (RIPA) total protein lysate (cat. No. P0013C, Beyotime, Inc., Shanghai, China) following the manufacturer’s instructions, then quantified using the bicinchoninic acid (BCA) kit (cat. No. BL524A, Biosharp, Inc., Anhui, China). Proteins from each sample were separated by electrophoresis on 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels and transferred onto polyvinylidene fluoride (PVDF) membranes (cat. No. HVLP00010, Millipore, Inc., Burlington, MA, USA). After incubation with WB-specific blocking solution [5% skimmed milk powder diluted in Tris-buffered saline with Tween 20 (TBST)], the PVDF membranes were separately incubated with primary antibodies at 4 °C overnight. After the blots were washed, they were incubated with horseradish peroxidase (HRP)-conjugated secondary antibody (1:1,000 dilution; cat. No. A0216, Beyotime, Inc. China) for 1.5 h and detected by a chemiluminescent imaging system (Bio-Rad, Hercules, CA, USA).
Enzyme-linked immunosorbent assay (ELISA)
The levels of IL-6 and TNF-α were measured in HK-2 cells using ELISA kits (cat. No. EH0201 and EH0302, FineTest, Inc., Wuhan, Hubei, China) according to the manufacturer's instructions. Optical density (OD) values were measured with a microplate reader (Multiskan FC, Thermo Fisher Scientific) in each well.
Acridine orange (AO) staining
The fluorescent dye AO was used to detect apoptotic cells. Cell staining with AO staining kit (cat. No. BB-4138B, BestBio, Inc., Shanghai, China) was performed according to the manufacturer’s instructions. Stained cells were visualized by a fluorescent microscope (IX71, Olympus, Shinjuku, Tokyo, Japan). Flow cytometric analysis was used to detect the acidic vesicular organelles (AVOs), which are a characteristic of autophagy.
Reactive oxygen species (ROS) assay
The ROS levels were measured using a ROS assay kit (cat. No. S0033S, Beyotime, Inc., China), following the manufacturer’s instructions. Stained cells were visualized by a fluorescent microscope (IX71, Olympus, Japan).
Immunofluorescence assay
We performed light chain 3 (LC3) immunofluorescence following standard immunofluorescence procedures. Briefly, cells were fixed with 4% methanol (cat. No. A0000700, Richjoint, Inc., Lioacheng, Shandong, China) followed by permeabilization with 0.1% Triton X-100 (cat. No. T8200, Solarbio, Inc., China) at room temperature. The cells were then blocked with 5% FBS (cat. No. 16140071, Gibco, Inc. USA), then incubated (4 °C) with the primary antibody, anti-LC3 (cat. No. ab51520, Abcam, Inc., Cambridge, MA, USA, 1:1,000) overnight. This procedure was followed by an incubation with the fluorescent Alexa Fluor 488® conjugated secondary anti-rabbit IgG (cat. No. 11008, Invitrogen, Inc., Carlsbad, CA, USA, 1:1,000) for 1 h at room temperature. After staining with 4’,6-diamidino-2-phenylindole (DAPI) and final washes with PBS, images were examined and captured using a fluorescence microscope (IX71, Olympus, Japan).
Dual luciferase assay
The relationship between miRNA and its predicted target sequences was detected using Dual-Luciferase® Reporter Assay System (cat. No. E1910, Promega, Inc., Madison, WI, USA), according to the manufacturer’s instructions. The data was read via the Multi-Mode Microplate Reader (SpectraMax i3x, Molecular Devices, San Jose, CA, USA).
Statistical analysis of data
Image analysis was conducted using ImageJ software (Fiji, ImageJ 1.53c; National Institutes of Health, Bethesda, MD, USA). The Origin software (version 6.1; https://www.originlab.com/) was used to perform statistical analysis and mapping. One-way analysis of variance (ANOVA) followed by Tukey’s post-hoc test was used to analyze differences between groups. Data were considered significantly different when P<0.05.
Results
LPS induced autophagy and silencing CYR61 alleviated autophagy
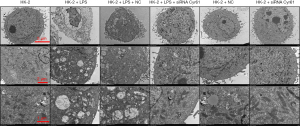
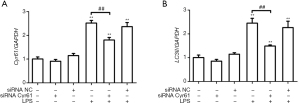
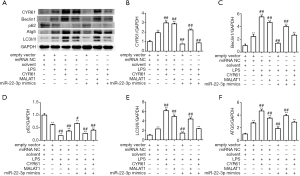
According to the pre-experimental results (Figure S1), We used 1 µg/mL LPS to incubate for 12 h to establish the HK-2 cell inflammation model. We used shRNA to silence CYR61. Among the 3 selected fragments, the knockdown effect of CCN1-siRNA1 was the most significant, so this sequence was selected for subsequent experiments (Figure S3). At the cellular level, as shown by the transmission electron microscopy results (Figure 1), in the control group and NC group, the nuclear membrane was clear, nucleolus was intact, organelles were relatively rich and evenly distributed, vacuoles in the cytoplasm were less and smaller, number of mitochondria was more, and the structure was clear. After LPS stimulation, a large number of vacuoles appeared, the nuclear membrane of the cells was wrinkled, edges were irregular, number of mitochondria decreased or the number of mitochondria was swollen or small, and the organelle content decreased. After silencing the CYR61 gene, the vacuoles in the cells became smaller and less, organelle content increased, and some mitochondria swelled. At the transcription and translation level, as shown in Figures 2,3, after LPS induction, the protein expression of CYR61 and LC3II increased significantly. After CYR61 was knocked down, the protein expression of CYR61 and LC3II showed a downward trend. These results suggested that autophagy occurs in the LPS-induced inflammation model, which requires the involvement of CYR61.

miR-22-3p and MALAT1 were the upstream regulators of CYR61
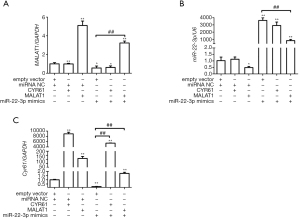
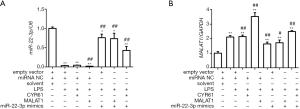
As described in the introduction, previous studies have shown that miR-22-3p inhibited AKI, while MALAT1 participated in AKI through recruitment of miR-22-3p. We speculated that CYR61 was their target protein. Firstly, the relationship between MALAT1, CYR61, and miR-22-3p was detected using dual-luciferase assay in HEK293 cells. As shown in Figure 4, miR-22-3p mimics significantly reduced the dual-luciferase activities of MALAT1(5440) and CYR61 3'-untranslated region (3'-UTR) but had no significant effect on MALAT1(3940). The inhibition of miR-22-3p significantly increased the dual-luciferase activities of MALAT1(5440) and CYR61 3'-UTR, but had no significant effect on MALAT1(3940). The results of RT-PCR showed that the MALAT1 overexpression vector significantly increased the intracellular CRY61 and MALAT1 expression in HK-2 cells, and significantly decreased the miR-22-3p expression. The miR-22-3p mimics significantly reduced the expression of CRY61 and MALAT1, while the CYR61 overexpression vector had no effect on MALAT1 and miR-22-3p. The results of WB showed that at the protein expression level, CYR61 overexpression vector and MALAT1 overexpression vector could significantly increase the expression of CRY61 in cells; miR-22-3p mimics could significantly reduce the expression of CRY61 in cells. The results are shown in Figures 5,6, revealing the regulatory relationship between CYR61 and miR-22-3p mimics and MALAT1: CYR61 was negatively regulated by miR-22-3p, while miR-22-3p and MALAT1 were negatively regulated with each other.


The effects of MALAT1 and miR-22-3p on apoptosis and autophagy
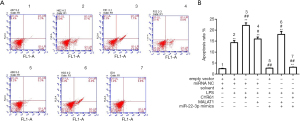
Based on the above results, the effects of MALAT1 and miR-22-3p on the apoptosis and autophagy of HK-2 cells were further examined. Annexin V-FITC/PI staining, AO staining, and flow cytometry showed that LPS, CYR61, and MALAT1 significantly increased cell apoptosis, while miR-22-3p mimics significantly reduced cell apoptosis, as shown in Figures 7,8. Immunofluorescence was used to detect the autophagy marker LC3, the results showed that LPS, CYR61 and MALAT1 could significantly increase the expression level of LC3 in cells, while miR-22-3p mimics reduced intracellular LC3 (Figure 9). Through the above experiments, we verified the effects of MALAT1 and miR-22-3p on the apoptosis and autophagy of HK-2 cells. It was shown that LPS, CYR61 overexpression, or MALAT1 overexpression induced cell apoptosis and autophagy, while miR-22-3p mimics had a protective effect on cells by down-regulating CYR61 and MALAT1.
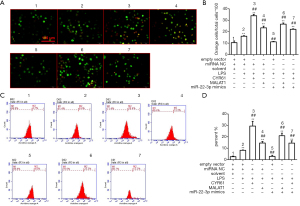
Molecular mechanisms by which MALAT1 and miR-22-3p regulate autophagy
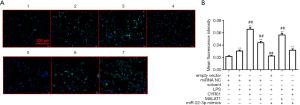
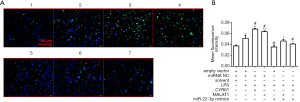
To further explore the molecular mechanism by which LPS, CYR61, and MALAT1 regulate autophagy, WB assay, and RT-qPCR were used to detect the regulation of autophagy-related proteins by LPS, CYR61, MALAT1, or miR-22-3p. As shown in Figure 10, LPS, CYR61 overexpression vector, and MALAT1 overexpression vector upregulated intracellular CYR61, Beclin1, Atg5, and LC3, and significantly downregulated p62 in the cell. The miR-22-3p mimics down-regulated the intracellular expressions of CYR61, Beclin1, Atg5, and LC3, and significantly up-regulated intracellular p62. As illustrated in Figure 11, LPS, CYR61, and MALAT1 significantly increased intracellular ROS levels, while miR-22-3p mimics significantly reduced ROS levels. These results further confirmed that LPS, CYR61, and MALAT1 induce autophagy by regulating autophagy-related proteins and intracellular ROS levels. The miR-22-3p mimics had a protective effect on cells by down-regulating CYR61 and MALAT1 (Figure 12).
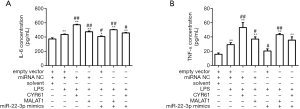
CYR61 and MALAT1 and miR-22-3p affect cellular inflammation by regulating TNF-α and IL-6
In order to detect the effects of LPS, CYR61, MALAT1, and miR-22-3p on inflammatory cytokines, we used an ELISA kit to detect intracellular IL-6 and TNF-α. As shown in Figure 13, LPS, CYR61, and MALAT1 significantly increased intracellular IL-6 and TNF-α, while MiR-22-3p mimics significantly down-regulated the expression of IL-6 and TNF-α in cells.
Discussion
In this study, we first established an LPS-induced inflammation model in HK-2 cells to identify apoptosis and autophagy in the condition of inflammation. Subsequently, shRNA knockdown of the CYR61 gene revealed LPS-induced autophagy reduction, which confirmed that CYR61 was a necessary condition for autophagy in the LPS-induced inflammation model. By using a dual-luciferase reporting system, RT-PCR, and WB assays, we identified the regulatory relationship between miR-22-3p, MALAT1, and CYR61. This revealed that miR-22-3p directly down-regulate CYR61, while miR-22-3p and MALAT1 negatively regulate each other. Furthermore, by using AO staining, immunofluorescence, WB assay, RT-PCR, and ELISA, we found that the overexpression of MALAT1 and CYR61 increased the levels of Beclin1, Atg5, LC3, ROS, IL-6, and TNF-α, significantly decreased the level of p62, and promoted apoptosis and autophagy. The miR-22-3p decreased the intracellular levels of CYR61, Beclin1, Atg5, LC3, ROS, IL-6, and TNF-α, up-regulated the intracellular level of p62, and inhibited apoptosis and autophagy. In conclusion, in the LPS-induced inflammation model, CYR61 was a necessary condition for autophagy, and miR-22-3p and MALAT1 were its upstream regulatory factors.
As shown in the results, the dual-luciferase assay revealed that miR-22-3p could bind to CYR61 3-'UTR, suggesting that miR-22-3p is an upstream regulator of CYR61. In HK-2 cells, miR-22-3p mimics significantly reduced the intracellular CYR61 level, which was consistent with the dual luciferin results. The expression level of MALAT1 did not change in the cells transfected with the CYR61 overexpression vector, and the expression level of CYR61 was up-regulated in the cells transfected with CYR61 overexpression vector, indicating that MALAT1 is also the upstream regulator of CYR61. In the level of transcription and protein expression, we found that miR-22-3p mimics and MALAT1 were negatively regulated, which is in line with the findings of Zhang et al. (22). In addition, Annexin V-FITC/PI staining, AO staining, and flow cytometry were used to detect the autophagy and apoptosis of inflammatory cells with overexpression of MALAT1 or CYR61 or with the addition of miR-22-3p mimic at the cell level. The results were also consistent with previous results.
The gene LC3 is a key gene in autophagy, its content is closely related to the activity of autophagy, and it is known as the marker molecule of autophagy (25). When autophagy occurs, LC3B gene expression on chromosome 16 will be up-regulated, and a large number of LC3B proteins will be translated and expressed (26). In the cytoplasm, LC3B protein is clipped by Atg4B protein to form LC3BI protein. By covalently binding PE on the autophagy membrane, LC3BI protein is eventually converted into a fat-soluble protein LC3BII, which plays a regulatory role in the extension and fusion of autophagosome membrane (27,28). Beclin1 is another key autophagy gene, which positively regulates autophagy to maintain the stability of the organism’s internal environment. The proteins encoded by it bind to cofactors (ATG 14L, UVRAG, and so on) to regulate the activity of vesicular protein sorting 34 (VPS-34) to form the Beclin 1-VPS34-VPS 15 complex, which promotes the formation of autophagosome membranes (29-31). The Atg5 gene plays an important role in the bending of the macrophage membrane and the recruitment of LC3 (32). Also, p62 can bind to ubiquitination vesicles and LC3 or their homologs, and is often used for autophagy detection (25). In this study, a variety of autophagy-related factors were detected, which ensured the credibility of our conclusion.
A previous study showed that TGF-β enhanced renal CYR61 after occurrences of unilateral ureteral obstruction (UUO) (16). Then, monocyte chemotaxis and macrophage infiltration were lead from activation of monocyte chemoattractant protein-1 by CYR61 (33). This evidence suggests that inhibition of CYR61 may prevent adverse consequences leading to irreversible AKI-CKD transition by delaying inflammation, intertubercle fibrosis, and apoptosis (34). Another study showed that after bilateral renal ischemic injury, CYR61 was rapidly stimulated in the proximal renal tubules and was excreted in urine within 3–6 h. Thus, urinary CYR61 might be used as a biomarker for AKI (9,35).
Current studies have shown that CYR61 plays an important role in lung disease, kidney disease, cardiovascular disease, liver disease, and other diseases (16,36-38). The mechanisms of its action are also varied, such as direct activation of toll-like receptor signal (39), activation of the PTEN/Akt/GSK3β/CyclinD1 signaling pathway (40), regulation of DCK and CTGF (41), inhibition of CD40 and its proteins related to non-canonical signaling (42), and so on. The complexity of its downstream pathways also indicates the complexity of its physiological and pathological effects, which is consistent with the phenomenon that the up-regulation of CYR61 may play different roles in the inflammation of different tissues.
In conclusion, CYR61 positively regulated autophagy of HK-2 cells in an inflammatory state and was negatively regulated by miR-22-3p, while miR-22-3p and MALAT1 were negatively regulated with each other.
Acknowledgments
Funding: This study was funded by National Natural Science Foundation of China (Project Number: 81860296); Guangxi Natural Science Foundation of China (Project number: 2017GXNSFDA198005 and 2018GXNSFAA281038); High-level talent scientific research project of the Affiliated Hospital of Youjiang Medical College for Nationalities (Project number: Y202011719).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://dx.doi.org/10.21037/tau-21-623
Data Sharing Statement: Available at https://dx.doi.org/10.21037/tau-21-623
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/tau-21-623). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Fortrie G, de Geus HRH, Betjes MGH. The aftermath of acute kidney injury: a narrative review of long-term mortality and renal function. Crit Care 2019;23:24. [Crossref] [PubMed]
- Zhu K, Song H, Zhang Z, et al. Acute kidney injury in solitary kidney patients after partial nephrectomy: incidence, risk factors and prediction. Transl Androl Urol 2020;9:1232-43. [Crossref] [PubMed]
- Bonomini M, Del Vecchio L, Sirolli V, et al. New Treatment Approaches for the Anemia of CKD. Am J Kidney Dis 2016;67:133-42. [Crossref] [PubMed]
- Sy J, Chen JLT, Kalantar-Zadeh K. New solutions to old problems—metabolic acidosis in chronic kidney disease. Ann Transl Med 2020;8:1256. [Crossref] [PubMed]
- Nath KA. Tubulointerstitial changes as a major determinant in the progression of renal damage. Am J Kidney Dis 1992;20:1-17. [Crossref] [PubMed]
- Shi M, Zeng X, Guo F, et al. Anti-Inflammatory Pyranochalcone Derivative Attenuates LPS-Induced Acute Kidney Injury via Inhibiting TLR4/NF-κB Pathway. Molecules 2017;22:1683. [Crossref]
- Su BC, Hsu PL, Mo FE. CCN1 triggers adaptive autophagy in cardiomyocytes to curb its apoptotic activities. J Cell Commun Signal 2020;14:93-100. [Crossref] [PubMed]
- Juric V, Chen CC, Lau LF. Fas-mediated apoptosis is regulated by the extracellular matrix protein CCN1 (CYR61) in vitro and in vivo. Mol Cell Biol 2009;29:3266-79. [Crossref] [PubMed]
- Muramatsu Y, Tsujie M, Kohda Y, et al. Early detection of cysteine rich protein 61 (CYR61, CCN1) in urine following renal ischemic reperfusion injury. Kidney Int 2002;62:1601-10. [Crossref] [PubMed]
- Lau LF, Lam SC. The CCN family of angiogenic regulators: the integrin connection. Exp Cell Res 1999;248:44-57. [Crossref] [PubMed]
- Wong M, Kireeva ML, Kolesnikova TV, et al. Cyr61, product of a growth factor-inducible immediate-early gene, regulates chondrogenesis in mouse limb bud mesenchymal cells. Dev Biol 1997;192:492-508. [Crossref] [PubMed]
- Choi J, Lin A, Shrier E, et al. Degradome products of the matricellular protein CCN1 as modulators of pathological angiogenesis in the retina. J Biol Chem 2013;288:23075-89. [Crossref] [PubMed]
- Kim KH, Chen CC, Monzon RI, et al. Matricellular protein CCN1 promotes regression of liver fibrosis through induction of cellular senescence in hepatic myofibroblasts. Mol Cell Biol 2013;33:2078-90. [Crossref] [PubMed]
- Meyer K, Hodwin B, Ramanujam D, et al. Essential Role for Premature Senescence of Myofibroblasts in Myocardial Fibrosis. J Am Coll Cardiol 2016;67:2018-28. [Crossref] [PubMed]
- Jun JI, Lau LF. The matricellular protein CCN1 induces fibroblast senescence and restricts fibrosis in cutaneous wound healing. Nat Cell Biol 2010;12:676-85. [Crossref] [PubMed]
- Lai CF, Chen YM, Chiang WC, et al. Cysteine-rich protein 61 plays a proinflammatory role in obstructive kidney fibrosis. PLoS One 2013;8:e56481 [Crossref] [PubMed]
- Li C, Zhao L, Wang Y, et al. Cysteine-rich protein 61, a specific ultra-early biomarker in kidney ischemia/reperfusion injury. Nephrology (Carlton) 2019;24:798-805. [Crossref] [PubMed]
- Wang X, Wang Y, Kong M, et al. MiR-22-3p suppresses sepsis-induced acute kidney injury by targeting PTEN. Biosci Rep 2020;40:BSR20200527 [Crossref] [PubMed]
- Wu Y, Huang C, Meng X, et al. Long Noncoding RNA MALAT1: Insights into its Biogenesis and Implications in Human Disease. Curr Pharm Des 2015;21:5017-28. [Crossref] [PubMed]
- Ding Y, Guo F, Zhu T, et al. Mechanism of long non-coding RNA MALAT1 in lipopolysaccharide-induced acute kidney injury is mediated by the miR-146a/NF-κB signaling pathway. Int J Mol Med 2018;41:446-54. [PubMed]
- Sherif IO, Al-Shaalan NH, Sabry D. Ginkgo Biloba Extract Alleviates Methotrexate-Induced Renal Injury: New Impact on PI3K/Akt/mTOR Signaling and MALAT1 Expression. Biomolecules 2019;9:691. [Crossref] [PubMed]
- Zhang Z, Li M, Zhang Z. lncRNA MALAT1 modulates oxaliplatin resistance of gastric cancer via sponging miR-22-3p. Onco Targets Ther 2020;13:1343-54. [Crossref] [PubMed]
- Okazaki K, Sakamoto K, Kikuchi R, et al. Mapping and characterization of FLC homologs and QTL analysis of flowering time in Brassica oleracea. Theor Appl Genet 2007;114:595-608. [Crossref] [PubMed]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001;25:402-8. [Crossref] [PubMed]
- Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell 2010;140:313-26. [Crossref] [PubMed]
- Amar N, Lustig G, Ichimura Y, et al. Two newly identified sites in the ubiquitin-like protein Atg8 are essential for autophagy. EMBO Rep 2006;7:635-42. [Crossref] [PubMed]
- Kabeya Y, Mizushima N, Yamamoto A, et al. LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J Cell Sci 2004;117:2805-12. [Crossref] [PubMed]
- Nakatogawa H, Ichimura Y, Ohsumi Y. Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell 2007;130:165-78. [Crossref] [PubMed]
- Funderburk SF, Wang QJ, Yue Z. The Beclin 1-VPS34 complex--at the crossroads of autophagy and beyond. Trends Cell Biol 2010;20:355-62. [Crossref] [PubMed]
- Thoresen SB, Pedersen NM, Liestøl K, et al. A phosphatidylinositol 3-kinase class III sub-complex containing VPS15, VPS34, Beclin 1, UVRAG and BIF-1 regulates cytokinesis and degradative endocytic traffic. Exp Cell Res 2010;316:3368-78. [Crossref] [PubMed]
- Kang R, Zeh HJ, Lotze MT, et al. The Beclin 1 network regulates autophagy and apoptosis. Cell Death Differ 2011;18:571-80. [Crossref] [PubMed]
- Geng J, Klionsky DJ. The Atg8 and Atg12 ubiquitin-like conjugation systems in macroautophagy. 'Protein modifications: beyond the usual suspects' review series. EMBO Rep 2008;9:859-64. [Crossref] [PubMed]
- Deshmane SL, Kremlev S, Amini S, et al. Monocyte chemoattractant protein-1 (MCP-1): an overview. J Interferon Cytokine Res 2009;29:313-26. [Crossref] [PubMed]
- Lai CF, Lin SL, Chiang WC, et al. Blockade of cysteine-rich protein 61 attenuates renal inflammation and fibrosis after ischemic kidney injury. Am J Physiol Renal Physiol 2014;307:F581-92. [Crossref] [PubMed]
- Edelstein CL. Biomarkers of acute kidney injury. Adv Chronic Kidney Dis 2008;15:222-34. [Crossref] [PubMed]
- Hsu PL, Chen JS, Wang CY, et al. Shear-Induced CCN1 Promotes Atheroprone Endothelial Phenotypes and Atherosclerosis. Circulation 2019;139:2877-91. [Crossref] [PubMed]
- Feng T, Meng J, Kou S, et al. CCN1-Induced Cellular Senescence Promotes Heart Regeneration. Circulation 2019;139:2495-8. [Crossref] [PubMed]
- Ju L, Sun Y, Xue H, et al. CCN1 promotes hepatic steatosis and inflammation in non-alcoholic steatohepatitis. Sci Rep 2020;10:3201. [Crossref] [PubMed]
- Jun JI, Lau LF. CCN1 is an opsonin for bacterial clearance and a direct activator of Toll-like receptor signaling. Nat Commun 2020;11:1242. [Crossref] [PubMed]
- Yan S, Liu H, Liu Z, et al. CCN1 stimulated the osteoblasts via PTEN/AKT/GSK3β/cyclinD1 signal pathway in Myeloma Bone Disease. Cancer Med 2020;9:737-44. [Crossref] [PubMed]
- Maity G, Ghosh A, Gupta V, et al. CYR61/CCN1 Regulates dCK and CTGF and Causes Gemcitabine-resistant Phenotype in Pancreatic Ductal Adenocarcinoma. Mol Cancer Ther 2019;18:788-800. [Crossref] [PubMed]
- Dang T, Meng X, Modak C, et al. Overexpression of CCN1 in Het1A cells attenuates bile-induced esophageal metaplasia through suppressing non-canonical NFκB activation. Cytokine 2019;116:61-9. [Crossref] [PubMed]


