
Bioinformatics analysis to screen DNA methylation-driven genes for prognosis of patients with bladder cancer
Introduction
Bladder cancer (BLCA) is the ninth leading cancer type globally, with 430,000 new cases diagnosed annually (1). In China, BLCA is 5 times more common among males relative to females (2), and rates of this cancer are rising. Owing to its high morbidity and mortality, BLCA is of key scientific interest. In 2015 alone in the USA, there were 74,000 urothelial carcinoma of the bladder (UCB) diagnoses and 16,000 BLCA-related deaths (3). The TNM system can grade tumors based upon cellular characteristics and degree of invasion (Tis-T4) and is graded according to its cellular characteristics. While recurrence is a common finding in those with non-muscle invasive bladder cancers (NMIBCs) (50–70%), the disease only becomes invasive in 0-15% of cases and the 5-year survival rate is currently 90% (4). Transurethral resection of bladder tumor (TURBT) is considered the standard of treatment for NIMBC. However, approximately 70% of patients will relapse, and it is estimated that 30% of patients will eventually need radical cystectomy due to the disease developing into muscle invasive bladder cancer (MIBC). There is no approved second-line treatment for patients who progress after first-line treatment (5). Even though surgery, radiotherapy, chemotherapy, and immunotherapy approaches to treating this cancer type have been designed, BLCA has remained a major therapeutic challenge owing to its complex pathogenesis.
Apart from environmental factors, genetic material mutation is one of the main BLCA-related risk factors. However, the molecular regulation mechanism of BLCA is still not entirely understood (6,7). The methylation of DNA is a key epigenetic process that is also linked to oncogenesis (8). Such methylation can regulate genomic stability, cellular differentiation, and many other processes, thereby potentially impacting cancer development and prognosis. Recently, increasing evidence has suggested that the onset of BLCA is a multigenic, multifactorial process, and there is thus a clear need for further study of the epigenetic basis for BLCA. Gene methylation can profoundly influence gene expression, an understanding of which is beneficial to BLCA diagnostic and prognostic evaluation. A total of 120 genes relating to the interaction between micro RNA (miRNA) and methylation have been discovered, and 11 important epigenetic interactions between 2 epigenome components have been found to be related to survival rate (9). A urine methylation biomarker classifier for BC monitoring has been developed. If cystoscopy were to be performed only on patients whose combined classifier results are positive, 36% of all potential cystoscopy could be prevented (10). Compared with paracancerous tissues, the expression of Dlg5 is reduced in most BLCA tissues, and the expression of Dlg5 is further down-regulated in patients with muscle invasive tumors. The hypermethylation of Dlg5 in bladder tumors is closely related to the silence of Dlg5 expression (11). It has been revealed that some special methylation-driven genes (MDGs) may be useful biomarkers for the diagnosis, therapy, and prognosis assessment of BLCA.
The Cancer Genome Atlas (TCGA) database is an open access research tool containing epigenetic and genetic information pertaining to a wide range of tumors that can be utilized for research purposes. At the present, differential methylation patterns and MDGs in several cancer subtypes have been identified through mining data from TCGA. However, the methylation patterns and the prognosis value of MDGs in BLCA are still unclear.
Herein, BLCA patient-related messenger RNA (mRNA) expression and methylation data were obtained from TCGA, and the methylation patterns and MDGs were identified using R language (http://www.R-project.org/). Then, MDGs and some clinical features were utilized for survival model construction, to assess the MDGs associated with BLCA prognosis, and explore correlations between DNA methylation and BLCA gene expression. This study has provided a rational foundation for personalized medicine of BLCA.
We present the following article in accordance with the TRIPOD reporting checklist (available at https://dx.doi.org/10.21037/tau-21-326).
Methods
Data collection and preprocessing
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). For this study, 399 samples of DNA methylation array data of TCGA-BLCA cohort were downloaded from TCGA (http://cancergenome.nih.gov/). Samples were randomized into a training set (n=272) and testing set (n=117). The DNA methylation array data was generated from the Illumina Infinium Human-Methylation450 Bead-Chip array (Illumina. San Diego, CA, USA). We used β values (between 0 and 1) to assess probe methylation levels. These β values were excluded when they mapped to mitochondrial, X, or Y chromosomes to prevent possible bias. In addition, the ~20% of samples with absentβ values were excluded. Furthermore, probes overlapping with repeat masker and single nucleotide polymorphisms (SNPs) from dbSNP v151 (National Center for Biotechnology Information (NCBI), Bethesda, MD, USA) with a minor allele frequency (MAF) >1% were also removed (12). The DNA methylation was analyzed in 399 samples at the regional and site levels, with probes independently mapped to 6 genomic sub-regions: transcription start site (TSS) 200 (TSS to 200 bp upstream of the TSS), TSS1500 (200–1,500 bp upstream of the TSS), 5’ UTR, 1st exon, gene body, and 3’UTR and intergenic regions (IGR). In addition,5’-C-phosphate-G-3’ (CpG) island, shore (0–2 kb from CpG islands), and shelf (2–4 kb from CpG islands) methylation patterns were also assessed.
Transcription-level data from the BLCA patients and complete clinical datasets were obtained from TCGA-BLCA (http://cancergenome.nih.gov/). A total of 395 samples of RNA-sequencing data were enrolled in this study. The DESeq2 (13) was employed for the expression of differentially expressed genes (DEGs) based on raw read counts. Those genes that had a false discovery rate (FDR) ≤0.05 and the absolute value of log2 fold change (FC) difference ≥1 (|log2 (FC)|≥1) were considered to be differentially expressed.
Consensus cluster analysis
Relevant variable CpG sites were extracted based upon a standard deviation (SD) threshold >0.1 in BLCA samples. Clustering analyses were then conducted with the RConcensusClusterPlus package based upon these variable sites and K-means clustering (14). The prespecified dataset was classified into k clusters via the algorithm. Stable clusters were obtained via 100 iterations with a mxk=20 parameter, with 80% of samples being used per analysis. The maximum number of clusters with a minimum of 90% clustering consensus was chosen, with a cumulative distribution function (CFD) and the delta area map being utilized to select optimal cluster numbers.
Survival analysis
We assessed BLCA subtype overall survival curves via the Kaplan-Meier (KM) approach as a function of methylation profiles. Differences between clusters were compared via log-rank tests. Associations between cluster biological and clinical findings were assessed via chi-squared tests. Survival analyses were performed with the survminer R package. The significance threshold was set at P<0.05.
DNA methylation analysis
Following preprocessing and imputations analyses, CpGβ values underwent further normalization with an Rβ mixed integer-quantile normalization (BMIQ) tool to control for type I and II probes (15). The R limma package was utilized for supervised differential methylation analysis. The CpG residues were deemed to be differentially methylated when the |log2 (FC)| of cluster1 and cluster2β value were ≥0.2, and the Benjamini-Hochberg (BH) adjusted P value ≤0.05. The gtrellis R package was utilized to generate circular 10 Mb sliding window plots for individual chromosomes assessing differentially methylated CpGs (dm-CpGs) with varying frequencies of methylation (16). Methylation frequencies per Mb pair for individual chromosomes were then determined by dividing the number of dm-CpGs per chromosome by chromosomal length in Mb based on the GRCh38 genome. The relative frequencies of hypomethylation and hypermethylation were additionally determined via a similar approach. When the hypermethylation to hypomethylation frequency ratio was ≥1.5 for a given chromosome, it was considered to be primarily hypomethylated.
Analysis of differential methylation regions
The DMRcate Bioconductor tool (http://www.bioconductor.org/packages/release/bioc/html/DMRcate.html) was utilized to analyze differential methylation regions (DMRs) (17). This tool first calculated the differential methylation of particular CpG residues with a limma-derived moderated t-statistic (18). After correcting for FDR, significant dm-CpGs regions were pooled when consecutive probes were in a 1 kb distance, with DMRs that had a minimum of 2dm-CpGs with an adjusted P value <0.01 within 1kb being incorporated into DMR analyses, with the Gviz Bioconductor package being used to plot the resultant DMRs (19).
Functional enrichment analyses
The GO and KEGG analyses were performed with the RclusterProfiler software (20). The significance threshold was set at P<0.05.
Protein-protein interaction (PPI) network
The PPI network of the aggregated DEGs was prepared and visualized with STRING (https://string-db.org/cgi/network.pl). A combined interaction score >0.4 was considered statistically significant.
Correlation analyses
The correlation between DNA methylation level and gene expression value was detected with Spearman’s correlation coefficient analysis. Correlation coefficient ≥0.3 and FDR ≤0.05 were considered statistically significant.
Establishment and verification of the prognostic prediction model
The hub DMRs and several clinical features were applied to a univariate Cox regression. Statistical significance was considered when P<0.1. Then, least absolute shrinkage and selection operator (LASSO) regression analysis was utilized to detect survival-related DMGs in the training set. Moreover, a multivariate Cox regression model was built to further identify the selected variates using ‘step’ function in R. Risk signatures were then generated through the linear combination of regression coefficient values from multivariate Cox regression model coefficient values and gene expression levels as follows:
Median risk score values were utilized to separate patients into low- and high-risk groups, and risk signature efficiency was evaluated via the KM approach and using time-dependent receiver operating characteristic (ROC)curves. The R rms package was used to construct a risk score-based overall survival (OS)-related nomogram. Calibration curves were generated, and C-index values were determined to assess nomogram efficiency. Furthermore, the prognostic predicated models also were validated in the testing set by constructing univariate Cox regression and a time-dependent ROC curve.
Statistical analysis
All statistical analyses were conducted using R software (version3.6.1). Univariate and LASSO Cox regression analyses were performed to construct and evaluate the prognostic predicated models using “glmnet” and “survival” packages of the R software. ROC curve analysis was performed to predict the OS of BLCA patients using the “survival ROC” package in the R. The OS between the two clusters was analyzed by Kaplan-Meier analysis with the log-rank test. A P value less than 0.05 was considered statistically significant.
Results
BLCA prognostic subtype methylation-based consensus clustering
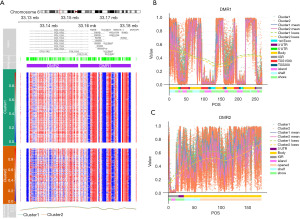
Methylation site consensus clustering was conducted to identify differential DNA methylation molecular subtypes for further prognostic analyses. The clustering result was relatively stable when k=2 as displayed by the cumulative distribution function (CDF) curve, despite that the delta area was significantly changed when k=2 (Figure 1A,1B). The results of consistent clustering indicated that the blue blocks were adjacent to each other on the white background when the clustering number was 2 (Figure 1C and Figure S1A-S1C). On the whole, all BLCA samples were divided into 2 clusters. Then, the effects of these 2 methylation subtypes on BLCA survival was investigated. The KM analyses suggested that the methylation consensus clustering-based prognosis of BLCA had insignificant differences between cluster1 and cluster2 (Figure 1D).

Differentialmethylation probes (DMPs) analysis
For the further analysis of DNA methylation in BLCA, we combined β values formal CpGs in relevant regions for cluster1 and cluster2. This analysis revealed a total 15,067 differentially methylated CpGs (dm-CpGs) between cluster1 and cluster2; of these, 14,189 exhibited hypermethylation and 878 exhibited hypomethylation (cluster2 vs. cluster1). As shown in Figure 2A, all dm-CpGs results from each autosomal chromosome shown on the out circle of the circus plot. In detail, chromosome 8 contained the highest, 290, methylation frequency (https://cdn.amegroups.cn/static/public/tau-21-326-1.xlsx). The most common DMP-related genes were: ERICH1-AS1, DLGAP2, CSMD1, MYOM2 and CTD-2281E23.2 (https://cdn.amegroups.cn/static/public/tau-21-326-1.xlsx). Normalization across chromosomes was achieved by dividing methylation by chromosomal size in Mb as a means of assessing differential methylation profiles. The results of this analysis revealed chromosome 17 as having the greatest mean frequency of hypomethylated sites while chromosome 6 had the highest mean frequency of hypermethylated sites (Figure 2B).

Then, the distribution of these dm-CpG sites were investigated. When assessing CpG residues in different locations, we found that the most apparent hyper- or hypo-methylated CpG sites were spread throughout the genome, while CpG islands were the most hypermethylated and the shelf regions exhibited the lowest degree of hypermethylation (Figure S2A). Based on the position relative to genes (1st Exon, 3’ UTR, 5’ UTR, body, TSS200, TSS1500and IGR), the distribution of methylated CpG sites indicated that the most hyper- and hypo- methylated CpG sites were located in the body (Figure S2B). The dm-CpG site distributions were then assessed based upon genetic and epigenetic annotations (Figure S2C). All of these results indicated that the differential CpG sites were mostly located in the CpG islands.
Differential methylated regions analysis
The DMRs comprise multiple consecutive methylated CpG sites with at least 2 dm-CpGs. Therefore, the effects of DMRs on gene expression and biological process are more significant than DMPs (21). In this study, cluster2 vs. cluster1, 146 DMRs across the genome in BLCA were identified. Chromosome 1 showed the highest number of DMRs (n=779) and chromosome 21 showed the lowest number of DMRs (n=37) (https://cdn.amegroups.cn/static/public/tau-21-326-1.xlsx). The distribution lengths of DMRs ranged from 149 bp to 26,103 bp. There were 2,172 long (>2,000 bp) DMRs. The number of dm-CpGs within DMRs ranged from 7 to 243, and there were 6 DMRs containing more than 100 dm-CpGs (https://cdn.amegroups.cn/static/public/tau-21-326-1.xlsx). As shown in Figure 3A, the DMR with the highest and second highest number of dm-CpGs (DMR1 and DMR2) revealed differential methylation patterns between cluster1 and cluster2 on chromosome 6. Several oncogenes, such as RXRB, RING1, and SLC39A7, were located in this region. The top 10 DMRs with CpG sites, DMR distribution, and related genes between cluster1 and cluster2 are presented in Figure S3. Moreover, the distribution of CpG on CpG island and gene regions in DMRs also displayed discernible methylation patterns between cluster1 and cluster2 (Figure 3B,3C and Figure S4).
Identification of DEGs
The DEGs were identified between cluster1 and cluster2. Volcano plots and heat mapping show the significantly changed genes with |log2 (FC)| ≥1 and FDR <0.05 in cluster2 compared with cluster1 (Figure 4A,4B). In total, 4,234 genes showed significantly differential expression, including 1,583 upregulated genes and 2,651 downregulated genes (https://cdn.amegroups.cn/static/public/tau-21-326-1.xlsx). In addition, https://cdn.amegroups.cn/static/public/tau-21-326-1.xlsx and Figure 4C display the details of the top 10 DEGs. Compared with cluster1, MTND1P23, SSTR5-AS1, GRM3, TMEN178A and SLC39A5 were significantly upregulated in cluster2, while RHOH, CD52, CD209, CD48 and IL10RA were significantly downregulated.
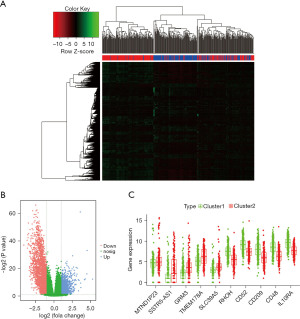
Next, the biological classification of DEGs was investigated. The KEGG pathway enrichment analyses revealed that upregulated genes predominantly participated in cancer-related pathways including the drug metabolism, retinol metabolism, chemical carcinogenesis, and cytochrome P450xenobiotic metabolism (Figure 5A). The downregulated genes of the KEGG pathways were mainly enriched in cytokine-cytokine receptor interactions, viral protein interactions with cytokines/cytokine receptors, and the chemokine signaling pathway (Figure 5B). The GO enrichment analysis showed that the upregulated genes were mainly involved in endocrine system development, steroid metabolic process, and glucuronosyltransferase activity (Figure 5C). The GO enrichment analysis of biological processes was enriched in adaptive immune response, positive regulation of cell activation, and leukocyte migration. Furthermore, cell component analysis indicated that these genes were enriched in the external side of plasma membrane and collagen-containing extracellular matrix. Cytokine activity and extracellular matrix structural constituent were the mainly enriched terms for molecular function (Figure 5D).

Selection and analysis of MDGs
To detect the regulatory effect of DNA methylation on gene expression, the relationship between dm-CpGs and DEGs was explored. The results demonstrated that 33 genes were negatively correlated with the corresponding CpGs (https://cdn.amegroups.cn/static/public/tau-21-326-1.xlsx), thus they were identified as MDGs for BLCA. The MDGs with the top 10 correlation coefficients are displayed in Figure 6A, including TBX3, IFFO1, SLITRK6, PSMB9, SRGN, TAP1, FAM78A, GPR68, EPHB6 and DHRS2.
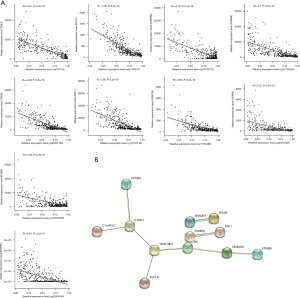
The PPI network was constructed to analyze the interaction of 33 MDGs. A PPI diagram with 12 node genes and 9 interaction is shown in Figure 6B.
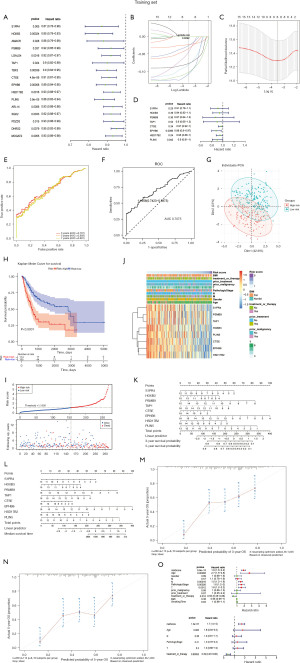
Construction of the prognosis prediction model in training set
To investigate the contribution of 33 MDGs to BLCA survival, a total 389 BLCA samples were distributed into a training set (n=272) and testing set (n=117), which were involved in identification and verification the prognostic prediction model (https://cdn.amegroups.cn/static/public/tau-21-326-1.xlsx). Thus, univariate Cox proportional hazard regression analyses were performed, revealing 16 MDGs including S1PR4, HOXB3, AMACR, PSMB9, LGALS4, TAP1, TBX3, CTSE, EPHB6, HSD17B2, PLIN5, ARL14, SGK2, PDZD3, DHRS2and MOGAT2 that were significantly associated with poor BLCA patient survival (Figure 7A and https://cdn.amegroups.cn/static/public/tau-21-326-1.xlsx). When the LASSO regression and multivariate Cox proportional hazard regression analyses were conducted in the training set, 8 BLCA-specific prognostic MDGs (S1PR4, HOXB3, PSMB9, TAP1, CTSE, EPHB6, HSD17B2and PLIN5) were identified (Figure 7B-7D, https://cdn.amegroups.cn/static/public/tau-21-326-1.xlsx). Then, the risk score was calculated according to the multivariate Cox proportional hazard regression model, and the BLCA samples were divided into high-risk score and low-risk score groups based on the optimistic threshold value (1.156) and area under the curve (AUC) value (0.7075) (Figure 7E,7F). The principal component analysis (PCA) result also demonstrated the obvious bias between high- and low-risk groups (Figure 7G). The KM plot exhibited significant differences between high- and low-risk groups (Figure 7H), where the high-risk core group showed a poor survival rate in BLCA patients (Figure 7H,7I). The heatmap showed a correlation between8 BLCA-specific prognostic MDGs and clinical characteristics, and the results showed the risk score significantly correlated to age, pathologic stage, and stage N/M (Figure 7J and https://cdn.amegroups.cn/static/public/tau-21-326-1.xlsx). Moreover, the nomograms of median survival time and 3/5-year survival time were constructed based on 8 BLCA-specific prognostic MDGs for BLCA (Figure 7K,7L). The calibration plots showed the predictive accuracy of predicated models, it reveled predicated survival rate approximately equivalent to actual survival (Figure 7M,7N). We further investigated whether clinical characteristics affected the accuracy of predicated models via univariate and multivariate Cox proportional hazard regression analysis, which showed that treatment or therapy and risk score acted as the risk factors in BLCA, and these risk factors were suitable for predicated models (Figure 7O, https://cdn.amegroups.cn/static/public/tau-21-326-1.xlsx). This result illustrated that this nomogram may offer potential clinical value with and without treatment or therapy.
Predictive model validation
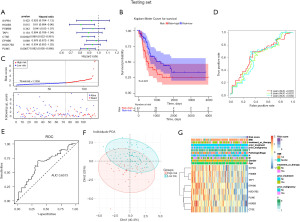
We further validated the prognostic predicated models via the testing set (Figure 8). Univariate Cox proportional hazard regression analyses in the testing set resultshowed3 BLCA-specific prognostic MDGs (HOXB3, CTSE, PLIN5) were identified as independent variates in BLCA (Figure 8A), and the ROC curve showed 3-year survival rate were predicated based on the univariate Cox model (AUC=0.6573) (Figure 8C). Based on the optimistic threshold of risk score in the training set, the testing set samples were divided into high- and low-risk score groups, which revealed that the high-risk score group showed poor survival rate in BLCA patients (Figure 8B,8E, https://cdn.amegroups.cn/static/public/tau-21-326-1.xlsx). The PCA plot also showed obvious bias between high- and low-risk score groups (Figure 8F). Furthermore, for the clinical characteristics involved in univariate Cox model analysis, it was revealed that risk score, pathologic stage, T/N/M stages, and smoking time acted as risk factors in BLCA (Figure 8G and https://cdn.amegroups.cn/static/public/tau-21-326-1.xlsx).
Discussion
The disease BLCA is the most common urological malignant tumor, and it has a poor prognosis (22). Although BLCA has attracted increasing attention, the mechanistic basis for BLCA remains to be clarified, and modern genome-wide DNA methylation analysis techniques represent a powerful means of analyzing patterns of BLCA-related methylation. The methylation of DNA plays critical roles in cancer biology by modulating gene expression, and DNA methylation pattern alterations can serve as available biomarkers for distinguishing tumors from normal samples. DNA methylation is a known factor leading to the development of BLCA. The earliest discovery that BLCA-related DNA methylation status may be related to the BLCA stage and level of potential genes (23). Catto et al. found that low-grade tumors have fewer changes in methylation sites compared with high-grade and invasive tumors (24). In low-grade non-invasive tumors, the frequency of DNA hypomethylation was higher than that of invasive tumors (25). At present, the diagnosis and prognostic analysis of BLCA can be performed based on DNA methylation status. Previous research has explored DNA methylation patterns in BLCA, and has revealed important genes and pathways that are dysregulated in BLCA (26). An article showed that detecting the methylation of genes such as POU4F2, PCDH17 and ONECUT2 through urine samples can efficiently detect BLCA (27). Ahlen et al. found that observing specific CpGs methylation status to assess CD4+ cell lineage can predict the prognosis of BLCA patients (28). In addition, Tian et al. identified that various prognostic subtypes of BLCA use dm-CpG sites (29). Several hub epigenetic MDGs had previously been investigated by Zhang et al. (30). Compared with previous studies, the current study was more in-depth, involving the assessment of differential methylation and DEGs. This study also examined correlations between these findings and BLCA patient survival and prognosis.
First, this study divided BLCA samples into 2 subtypes according to DNA methylation using consensus clustering. Despite that there were no significant differences in the OS between the 2 subgroups, 15,067 specific hyper- and hypo- methylation CpG sites were found. Specifically, dm-CpGs were found on all chromosomes in BLCA. The CpG islands and promoters had more differential CpG sites compared with other regions. Several hypomethylation CpG-related genes had a high frequency in BLCA, including PTPRN2, PRDM16, and NTM; these genes have been reported as oncogenes, involved in malignant biological behaviors (31-33). The gene PTPRN2 is upregulated in highly metastatic breast cancer cells, and its increased expression is linked to metastatic recurrence in humans (32). In patient tissues and in vitro models after treatment with estrogen and progesterone, NTM in leiomyomas was increased compared with in myometrium. Expression down-regulation occurred after ulipristal acetate (UPA) treatment, but not after fulvestrant exposure (31). This study was the first to suggest that these hypomethylated genes may regulate BLCA development.
In addition, this study screened out 4,234 DEGs between cluster2 and cluster1. Although all samples had been obtained from BLCA patients, the genes were differentially expressed, indicating that the gene expression was regulated by methylation pattern in BLCA (34). We then investigated the potential biological functions, and revealed that the most conspicuous pathways were cancer- and inflammation-related. Cancer onset and progression is closely linked to inflammation, although many tumors can evade the immune system. Such inflammation is related to chemokines, prostaglandins, and cytokines, which have been shown to down-regulate the activity of cytochrome P450 (CYP) enzymes (35). In total, 46 immune-related genes that were differentially expressed were linked to papillary carcinoma (PTC) patient clinical outcomes. Functional enrichment analyses revealed these genes to be involved in the cytokine-cytokine receptor interaction KEGG pathway (36).
Furthermore, 33 MDGs were identified by using Spearman’s correlation coefficient analysis. Following univariate Cox and LASSO regression analyses and stepwise regression model establishment with MDGs and common clinical features,8 epigenetic hub MDGs were identified, of which S1PR4, HOXB3, PSMB9, TAP1, CTSE, EPHB6, HSD17B2, and PLIN5were independent prognostic factors of BLCA. Using the risk model, BLCA patients could be stratified into high- and low-risk subgroups. In this study, the high-risk subgroup had higher risks of OS. Therefore, this predictive signature may facilitate the assessment of risk score of BLCA and construction of appropriate clinical follow-up plans accordingly. The AUC was 0.7075 by ROC analysis in the training set, suggesting the predictive accuracy was relatively ideal in this study. Finally, a nomogram including age, pathologic stage, treatment, and 8 hub MDGs was constructed to predict individual prognosis. Besides, 3 hub MDGs were identified as independent variates in the testing set. TheHOXB3, CTSE, and PLIN5 genes were identified as the specific prognostic MDGs. The calibration curves for 3-/5-/7-year survival rates were effectively predicted in both training and testing set BLCA patients. Thus, this nomogram may provide an accurate prognosis prediction for BLCA.
Conclusions
In summary, this study discovered differential methylation patterns and MDGs in BLCA patients, and found them to be promising biomarkers for the early diagnostic and prognostic assessment of BLCA patients.
Acknowledgments
Funding: This study was supported by Shenzhen Baoan Shiyan People’s Hospital Funding (2020SY07).
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://dx.doi.org/10.21037/tau-21-326
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/tau-21-326). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Cumberbatch MG, Noon APEAU Young Academic Urologists—Urothelial Cancer Working party. Epidemiology, aetiology and screening of bladder cancer. Transl Androl Urol 2019;8:5-11. [Crossref] [PubMed]
- Li K, Lin TChinese Bladder Cancer Consortium, et al. Current status of diagnosis and treatment of bladder cancer in China - Analyses of Chinese Bladder Cancer Consortium database. Asian J Urol 2015;2:63-9. [Crossref] [PubMed]
- Dobruch J, Daneshmand S, Fisch M, et al. Gender and Bladder Cancer: A Collaborative Review of Etiology, Biology, and Outcomes. Eur Urol 2016;69:300-10. [Crossref] [PubMed]
- Suh J, Yoo S. Role of immunotherapy in Bacillus Calmette-Guérin unresponsive: non-muscle invasive bladder cancer. Transl Cancer Res 2020;9:6537-45. [Crossref]
- Vasekar M, Degraff D, Joshi M. Immunotherapy in Bladder Cancer. Curr Mol Pharmacol 2016;9:242-51. [Crossref] [PubMed]
- Porten SP. Epigenetic Alterations in Bladder Cancer. Curr Urol Rep 2018;19:102. [Crossref] [PubMed]
- Cumberbatch MGK, Jubber I, Black PC, et al. Epidemiology of Bladder Cancer: A Systematic Review and Contemporary Update of Risk Factors in 2018. Eur Urol 2018;74:784-95. [Crossref] [PubMed]
- Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell 2007;128:669-81. [Crossref] [PubMed]
- Shivakumar M, Lee Y, Bang L, et al. Identification of epigenetic interactions between miRNA and DNA methylation associated with gene expression as potential prognostic markers in bladder cancer. BMC Med Genomics 2017;10:30. [Crossref] [PubMed]
- van der Heijden AG, Mengual L, Ingelmo-Torres M, et al. Urine cell-based DNA methylation classifier for monitoring bladder cancer. Clin Epigenetics 2018;10:71. [Crossref] [PubMed]
- Zhou Z, Guo Y, Liu Y, et al. Methylation-mediated silencing of Dlg5 facilitates bladder cancer metastasis. Exp Cell Res 2015;331:399-407. [Crossref] [PubMed]
- Zhou W, Laird PW, Shen H. Comprehensive characterization, annotation and innovative use of Infinium DNA methylation BeadChip probes. Nucleic Acids Res 2017;45:e22 [PubMed]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [Crossref] [PubMed]
- Wilkerson MD, Hayes DN. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics 2010;26:1572-3. [Crossref] [PubMed]
- Teschendorff AE, Marabita F, Lechner M, et al. A beta-mixture quantile normalization method for correcting probe design bias in Illumina Infinium 450 k DNA methylation data. Bioinformatics 2013;29:189-96. [Crossref] [PubMed]
- Gu Z, Eils R, Schlesner M. gtrellis: an R/Bioconductor package for making genome-level Trellis graphics. BMC Bioinformatics 2016;17:169. [Crossref] [PubMed]
- Peters TJ, Buckley MJ, Statham AL, et al. De novo identification of differentially methylated regions in the human genome. Epigenetics Chromatin 2015;8:6. [Crossref] [PubMed]
- Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43:e47 [Crossref] [PubMed]
- Hahne F, Ivanek R. Visualizing Genomic Data Using Gviz and Bioconductor. Methods Mol Biol 2016;1418:335-51. [Crossref] [PubMed]
- Yu G, Wang LG, Han Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 2012;16:284-7. [Crossref] [PubMed]
- Bert SA, Robinson MD, Strbenac D, et al. Regional activation of the cancer genome by long-range epigenetic remodeling. Cancer Cell 2013;23:9-22. [Crossref] [PubMed]
- Black AJ, Black PC. Variant histology in bladder cancer: diagnostic and clinical implications. Transl Cancer Res 2020;9:6565-75. [Crossref]
- Martinez VG, Munera-Maravilla E, Bernardini A, et al. Epigenetics of Bladder Cancer: Where Biomarkers and Therapeutic Targets Meet. Front Genet 2019;10:1125. [Crossref] [PubMed]
- Catto JW, Azzouzi AR, Rehman I, et al. Promoter hypermethylation is associated with tumor location, stage, and subsequent progression in transitional cell carcinoma. J Clin Oncol 2005;23:2903-10. [Crossref] [PubMed]
- Wolff EM, Chihara Y, Pan F, et al. Unique DNA methylation patterns distinguish noninvasive and invasive urothelial cancers and establish an epigenetic field defect in premalignant tissue. Cancer Res 2010;70:8169-78. [Crossref] [PubMed]
- Olkhov-Mitsel E, Savio AJ, Kron KJ, et al. Epigenome-Wide DNA Methylation Profiling Identifies Differential Methylation Biomarkers in High-Grade Bladder Cancer. Transl Oncol 2017;10:168-77. [Crossref] [PubMed]
- Wu Y, Jiang G, Zhang N, et al. HOXA9, PCDH17, POU4F2, and ONECUT2 as a Urinary Biomarker Combination for the Detection of Bladder Cancer in Chinese Patients with Hematuria. Eur Urol Focus 2020;6:284-91. [Crossref] [PubMed]
- Ahlén Bergman E, Hartana CA, Johansson M, et al. Increased CD4+ T cell lineage commitment determined by CpG methylation correlates with better prognosis in urinary bladder cancer patients. Clin Epigenetics 2018;10:102. [Crossref] [PubMed]
- Tian Z, Meng L, Long X, et al. DNA methylation-based classification and identification of bladder cancer prognosis-associated subgroups. Cancer Cell Int 2020;20:255. [Crossref] [PubMed]
- Zhang C, Shen K, Zheng Y, et al. Genome-wide screening of abberant methylated drivers combined with relative risk loci in bladder cancer. Cancer Med 2020;9:768-82. [Crossref] [PubMed]
- Parikh TP, Malik M, Britten J, et al. Steroid hormones and hormone antagonists regulate the neural marker neurotrimin in uterine leiomyoma. Fertil Steril 2020;113:176-86. [Crossref] [PubMed]
- Sengelaub CA, Navrazhina K, Ross JB, et al. PTPRN2 and PLCβ1 promote metastatic breast cancer cell migration through PI(4,5)P2-dependent actin remodeling. EMBO J 2016;35:62-76. [Crossref] [PubMed]
- Wang W, Ishibashi J, Trefely S, et al. A PRDM16-Driven Metabolic Signal from Adipocytes Regulates Precursor Cell Fate. Cell Metab 2019;30:174-189.e5. [Crossref] [PubMed]
- Li HT, Duymich CE, Weisenberger DJ, et al. Genetic and Epigenetic Alterations in Bladder Cancer. Int Neurourol J 2016;20:S84-94. [Crossref] [PubMed]
- Harvey RD, Morgan ET. Cancer, inflammation, and therapy: effects on cytochrome p450-mediated drug metabolism and implications for novel immunotherapeutic agents. Clin Pharmacol Ther 2014;96:449-57. [Crossref] [PubMed]
- Lin P, Guo YN, Shi L, et al. Development of a prognostic index based on an immunogenomic landscape analysis of papillary thyroid cancer. Aging (Albany NY) 2019;11:480-500. [Crossref] [PubMed]


