
TLN2 functions as a tumor suppressor in clear cell renal cell carcinoma via inactivation of the Wnt/β-catenin signaling pathway
Introduction
Renal cell carcinoma (RCC) arises from the renal tubular epithelial cells and ranks second among all types of urological malignancies (1). Data from recent studies have shown that the incidence of RCC has increased significantly compared with the past. Epidemiological studies have shown that RCC accounts for approximately 3% of new malignancies in adults each year, and its incidence has been on the rise over the past two decades (2). Clear cell RCC (ccRCC) is the most common and most malignant histological type of RCC, accounting for 70–85% of the incidence of RCC (3). At present, radical nephrectomy is the main treatment modality for RCC in clinical practice, but there is still a lack of proven methods for treating patients with advanced renal cancer and those with recurrence and metastasis after renal cancer surgery (4,5). Due to the lack of reliable clinical diagnostic biomarkers and typical early clinical symptoms, approximately more than 25% of RCC patients are already metastatic at the time of diagnosis, which makes simple surgical treatment difficult and shortens the survival time of patients (6). Compared with tumors such as lung and breast cancer, molecular markers for the diagnosis and metastatic recurrence of ccRCC are still poorly understood. Therefore, finding tumor markers and therapeutic targets with high sensitivity and specificity is crucial to improve the survival of ccRCC patients and is currently a hot research topic.
Past studies have found that a variety of factors in the body promote the malignancy of ccRCC, including aberrant expressions of growth factors, inactivation of tumor suppressors and activation of oncogenes (7-9). With extensive research, a large number of genes or signaling pathways that are closely related to the development and progression of RCC have been identified, including von Hippel-Lindau tumor suppressor (VHL), and vascular endothelial growth factor (VEGF) (10,11). Although several targeted therapeutic drugs for RCC have been introduced in recent years, their effects remain limited because of problems such as drug resistance. Therefore, it is necessary to carry out further basic experiments for in-depth research of the changes of tumor suppressors/oncogenes to explore the pathogenesis of ccRCC. Only by fully understanding the biological behavior of ccRCC can we find more novel targets and effective therapeutic drugs.
Wnt/β-catenin signaling is one of the key cascades that regulate development and stemness and has also been tightly associated with carcinogenesis (12). Numerous studies have focused on this signaling pathway in a variety of human cancers, including ccRCC (13). Talin is a macromolecular cytoskeleton protein located on the extracellular matrix (ECM) that can bind to a variety of adhesion molecules (e.g., integrin, actin, and adhesion kinase) (14). Talin is also an important component of focal adhesion (FA) plaque, which can activate the integrin/FA kinase signaling pathway by activating integrin, thereby affecting tumor metastasis and recurrence (15). At the same time, it can couple integrin to F-actin (16). Studies have shown that humans have two Talin genes, TLN1 and TLN2, which encode Talin 1 and Talin 2 proteins respectively. TLN2 has 74% homology with Talin 1. Talin1 can regulate FA dynamics, cell migration and cell invasion (17,18). Recent studies have shown that TLN2 knockdown can inhibit the migration of liver cancer cells (19), but the mechanism of TLN2 in ccRCC invasion and metastasis is still unclear. We present the following article in accordance with the ARRIVE reporting checklist (available at https://tau.amegroups.com/article/view/10.21037/tau-21-914/rc).
Methods
Bioinformatics analysis
TLN2 clinical information and expression data of ccRCC were downloaded from the GEPIA website (http://gepia.cancer-pku.cn/). Data from 523 ccRCC samples and 100 adjacent normal tissues were collectively analyzed. Differences of TLN2 levels between ccRCC and normal tissues were compared and the survival rates of ccRCC patients were analyzed.
Tissue samples
Sixty-two patients with ccRCC who received nephron-sparing or radical nephrectomy were enrolled in this study. No patient had received any adjuvant preoperative anti-cancer therapies. The tumor tissue samples and adjacent normal renal tissues were obtained during surgery with informed consent from the patients/patients’ families and the tissue samples were instantly frozen in liquid nitrogen for further analysis. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by ethics committee of Quanzhou First Hospital Affiliated to Fujian Medical University (No.: QZFH.FJMU.2018.175) and informed consent was taken from all the patients.
Immunohistochemistry (IHC)
Paraffin-embedded tissues were sliced into 4-µm sections. After deparaffination and then rehydration, the sections underwent microwave-stimulated antigen retrieval. Suppression of endogenous peroxidase activity was carried out with 3% hydrogen peroxide, followed by incubation of primary TLN2 (sc365460; 1:200; Santa Cruz, Dallas, TX, USA) antibody overnight at 4 °C. Subsequently, the slides were incubated with horseradish peroxide (HRP)-conjugated secondary antibody. After visualization signal development with DAB solution, the sections were counterstained with hematoxylin. A microscope (BX51; Olympus Corporation, Tokyo, Japan) was used to observe the results.
Cell lines and cell culture
Caki-1, A-498, 786-O, ACHN and HK-2 were obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). All cell lines were maintained in DMEM containing 10% fetal bovine serum (FBS) in a humidified incubator with 5% CO2 at 37 °C.
Cell transfection
TLN2 overexpression was achieved by transfections of TLN2 expression plasmids. The TLN2 expression plasmids and control vectors were purchased from Gene Pharma Company (Shanghai, China). Full length TLN2 cDNA was subcloned into an expression vector (pcDNA3.1/+) using the primer sequences: Forward: 5’-CG-GAATTC-ATGGTGGCCCTGTCCTTAAAG-3’, Reverse:5’-CCC-AAGCTT-TTAGCCCTCATCTTCCCTCAG-3’. Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) was used to transfect the TLN2 overexpression plasmid and control vector (an empty pcDNA3.1/+ plasmid) into ccRCC cells.
RT-qPCR assays
Total RNA isolation from targeted cells or tissues was performed by Trizol reagent (Invitrogen, USA). cDNAs were obtained by reverse transcription using Superscript III reverse transcriptase (Invitrogen). The expression of mRNA was examined using SYBR green PCR Master Mix (Takara) on the ABI 7900 Prism HT (Applied Biosystems). GAPDH was an internal control. The data were analyzed by a 2-ΔΔCT method. TLN-2: sense, 5’-CTG AGG CTC TTT TCA CAG CA-3’ and anti-sense, 5’-CTC ATC TCA TCT GCC AAG CA-3’; sense: 5’-CAT GAG AAG TAT GAC AAC AGC CT-3’ and anti-sense, 5’- AGT CCT TCC ACG ATA CCA AAG T-3’.
CCK-8 assay
CCK-8 assay (CCK-8, Dojindo, Japan) was performed to measure cell proliferation ability. The transfected Caki-1 and 786-O cells were seeded into a 96-well plate and incubated for 1, 2, 3, and 4 days. After incubation for the indicated times, CCK-8 solution was added. Cell viability was estimated by measuring the absorbance at 450 nm under a microplate reader after incubation for 2 h at 37 °C.
Colony formation
ccRCC cells (5×102/well) transfected with TLN2 expression plasmids or controls were seeded into a 6-well plate, and cultured for 14 days. The medium was changed every 3 or 4 days. After cell colonies had been formed, the cells were fixed with paraformaldehyde and stained with crystal violet. Visible cell colonies (≥50 cells were considered a colony) were photographed and counted under a microscope. Colony formation rate (%) = (colony numbers/inoculated cell numbers) ×100%.
Apoptosis assay
After transfection, the Caki-1 and 786-O cells were harvested, washed, and resuspended in ice-cold phosphate-buffered saline. Next, they were stained with propidium iodide (PI) and Annexin V-FITC (BD Biosciences, Lake Franklin, NJ, USA) and incubated at room temperature in the dark for 15 min. The data were analyzed using a FACS Calibur flow cytometer (BD Biosciences).
Cell cycle analysis
For cell cycle analysis, the transfected cells were fixed with ice-cold 70% ethanol, then stained with PI at 37 °C for 30 min in the dark. After staining, FACS Calibur (BD Biosciences) was utilized to analyze cell cycle distribution and the data were analyzed with the help of FlowJo software (BD Biosciences).
Xenograft tumor and metastasis assay
A total of 20 BALB/c nude mice (aged 4–6 weeks; female mice were selected due to their docility; 15–20 g) were used to establish in vivo xenograft models. The mice were fed under the same conditions and randomly divided into two groups. The animal experiment was approved by the hospital’s animal ethics committee. To establish a tumor growth model, Caki-1-control or Caki-1-TLN2 cells (5×106) were injected subcutaneously into the right flank of each mouse. The tumors were measured every 4 days, and 4 weeks later, the mice were humanely killed before the tumors were photographed and weighed. All efforts were made to minimize suffering. The tumor volume was calculated by length × width2 × 0.5. For the lung metastases assay, ccRCC cells were injected into the tail vein of the mice, and 6 weeks after the inoculation, the animals were killed humanely, and the bilateral lung tissues were excised and photographed, before undergoing HE staining for histological validation. Animal experiments were performed under a project license (No.: QZFH.FJMU.2018.a261) granted by the Animal Care and Use Committee of Quanzhou First Hospital Affiliated to Fujian Medical University, in compliance with institutional and national regulations for the care and use of animals. A protocol was prepared before the study without registration.
Wound healing
Caki-1 and 786-O cells were inoculated into a 6-well plate and incubated in DMEM with 10% FBS for 24 h to 100% confluence. The plate was scraped with the tip of a 100-µL plastic pipette to create a cell-free wound. The cells were cultured for another 24 h before the widths of the wounds at indicated time points (0 and 24 h) were examined using an Olympus BX51 microscope (Olympus, Tokyo, Japan).
Transwell assay
Transwell assays were conducted with a transwell chamber (8-µm pore; BD Biosciences) with/without Matrigel. For invasion assays, the chambers were first coated with Matrigel, and transfected cells were inoculated in the top chamber in serum-free DMEM, while for the migration assay, the chambers were not coated with Matrigel. In the meantime, the lower chambers were supplemented with culture medium containing 10% FBS and cells were incubated at 37 °C for 24 h. After being fixed with paraformaldehyde and stained with 0.1% crystal violet, the penetrating cells in five random fields were imaged and quantified with a microscope.
Western blot
Total proteins from cultured cells and targeted tissue samples were lysed in RIPA buffer (Thermo Scientific, Rockford, IL, USA). The concentrations of total proteins were detected by the BCA method, followed by separation by 10% SDS-PAGE. The proteins were then transferred to PVDF membranes (Millipore, Billerica, MA, USA). The membranes were blocked in 5% nonfat milk for 1 h at room temperature and incubated with primary antibodies at 4 °C overnight. Subsequently, the membrane was incubated with HRP-conjugated, secondary antibody (1:2000; Abcam, Cambridge, MA, USA). Protein bands were visualized with an enhanced chemiluminescence detection system (Pierce, Rockford, IL, USA). The primary antibodies against TLN2, β-catenin, c-Myc, cyclin D1, E-cadherin, N-cadherin, vimentin and GAPDH were obtained from Santa Cruz and the primary antibodies against cleaved caspase-3 and pro-caspase-3 were obtained from Abcam. GAPDH served as the loading control.
Statistical analysis
SPSS 23.0 software (IBM, USA) was used. For comparisons, one-way ANOVA, Student’s t-test and Chi-square were used as appropriate. The risk factors for the prognosis of ccRCC patients were determined by univariable and multivariable Cox regression analyses. P<0.05 was considered as a statistically significant difference.
Results
Downregulation of TLN2 in ccRCC clinical samples and cells
To verify the exact role of TLN2 in ccRCC, its levels in ccRCC tissues and cells were examined. Firstly, data from TCGA database showed that TLN2 expression in ccRCC tissues was prominently decreased compared with normal kidney tissue samples (Figure 1A). To further confirm the downregulation of TLN2 in ccRCC tissues, the TLN2 levels in 62 ccRCC and paired pericarcinomatous tissues were also detected. The RT-qPCR results indicated that TLN2 was significantly decreased in ccRCC tissues compared with the pericarcinomatous tissues (Figure 1B). The decreased level of TLN2 in ccRCC tissues was also confirmed by western blot (Figure 1C). Furthermore, the IHC results revealed that TLN2 was mainly expressed in the cytoplasm and TLN2 protein levels were obviously lower than in matched adjacent normal tissues (Figure 1D). In addition, the ccRCC cell lines (Caki-1, A-498, 786-O and ACHN) also showed lower TLN2 levels than HK-2 (Figure 1E,1F).
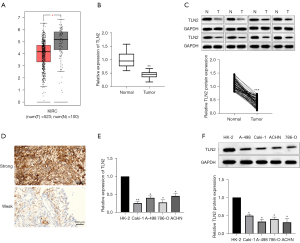
Effect of low expression of TLN2 on survival and correlation with disease progression of ccRCC patients
We further searched the TCGA database to determine the underlying prognostic significance of TLN2 in ccRCC patients. As shown in Figure 2, the patients with low TLN2 expressions showed poorer overall survival (OS) (Figure 2A) and disease-free survival (Figure 2B) than patients with high TLN2 expression. Subsequently, the relationship between TLN2 expression and the clinicopathological features of ccRCC patients was confirmed by Chi-square test. The enrolled patients were assigned to high-TLN2 and low-TLN2 groups according to the median TLN2 level. Results indicated that TLN2 level correlated remarkably with TNM stage and lymph node metastasis, while no significant correlation was found between TLN2 level and age, sex, and tumor size (Table 1). These results suggested that TLN2 participates in ccRCC progression.
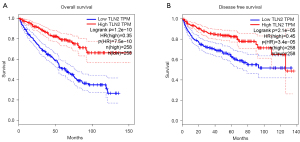
Table 1
| Clinicopathological features | Cases (n=62) | TLN2 expression | P value | |
|---|---|---|---|---|
| High (n=31) | Low (n=31) | |||
| Age (years) | 1 | |||
| >60 | 28 | 14 | 14 | |
| ≤60 | 34 | 17 | 17 | |
| Sex | 0.0944 | |||
| Male | 36 | 20 | 16 | |
| Female | 26 | 11 | 15 | |
| Tumor size (cm) | 0.2031 | |||
| ≥5.0 | 33 | 14 | 19 | |
| <5.0 | 29 | 17 | 12 | |
| TNM stage | 0.0023* | |||
| I–II | 30 | 21 | 9 | |
| III–IV | 32 | 10 | 22 | |
| Fuhrman grade | 0.2003 | |||
| G1–2 | 35 | 20 | 15 | |
| G3–4 | 27 | 11 | 16 | |
| Laterality | 0.2031 | |||
| Right | 33 | 19 | 14 | |
| Left | 29 | 12 | 17 | |
| Lymph node metastasis | 0.0110* | |||
| Present | 32 | 21 | 11 | |
| Absent | 30 | 10 | 20 | |
*, statistically significant. TNM, tumor-node-metastasis.
Cox regression analysis was then performed to further explore the prognostic potential of TLN2 in ccRCC patients. As demonstrated by univariate analysis, TLN2 expression [hazard ratio (HR) =5.092, P<0.001], tumor size (HR =0.402, P=0.027), TNM stage (HR =2.540, P=0.013) and lymph node metastasis (HR =4.538, P=0.001) were prominently associated with ccRCC OS (Table 2). Multivariate analysis indicated that TLN2 was an independent prognostic factor for ccRCC (Table 2). Taken together, TLN2 may be a promising biomarker for ccRCC progression and prognosis.
Table 2
| Variable | Univariate analysis | Multivariate analysis | |||
|---|---|---|---|---|---|
| HR (95% CI) | P value | HR (95% CI) | P value | ||
| Age (>60 vs. ≤60 years) | 1.223 (0.582–2.573) | 0.595 | 0.995 (0.413–2.395) | 0.991 | |
| Sex (Male vs. Female) | 1.078 (0.522–2.224) | 0.840 | 0.768 (0.337–1.750) | 0.530 | |
| Tumor size (≥5.0 vs. <5.0 cm) | 0.402 (0.179–0.904) | 0.027* | 0.730 (0.289–1.844) | 0.506 | |
| TNM stage (I–II vs. III–IV) | 2.540 (1.218–5.293) | 0.013* | 3.301 (1.313–8.297) | 0.011* | |
| Fuhrman grade (G1–2 vs. G3–4) | 1.893 (0.877–4.088) | 0.104 | 1.151 (0.495–2.675) | 0.744 | |
| Laterality (Right vs. Left) | 0.935 (0.446–1.961) | 0.859 | 0.978 (0.425–2.250) | 0.959 | |
| Lymph node metastasis (Present vs. Absent) | 4.538 (1.851–11.125) | 0.001* | 3.352 (1.141–9.849) | 0.028* | |
| TLN2 (High vs. Low) | 5.092 (2.155–12.031) | 0.000* | 2.953 (1.093–7.977) | 0.033* | |
*, P<0.05. HR, hazard ratio; CI, confidence interval.
Effect of TLN2 overexpression on growth and colony formation of ccRCC cells
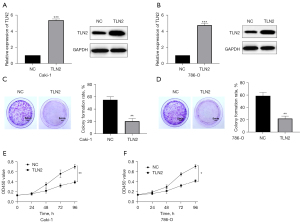
We overexpressed TLN2 expression in ccRCC cells to explore the underlying mechanism of TLN2 in inhibiting ccRCC. RT-qPCR and western blot were performed to determine the TLN2 expression after infection. As shown in Figure 3A,3B, the results demonstrated that the TLN2 vector efficiently increased TLN2 expression in ccRCC cells. To determine the effects of TLN2 on ccRCC cell viability, we performed colony formation and CCK-8 assays. The colony formation assays showed that TLN2 upregulation significantly suppressed ccRCC cell proliferation (Figure 3C,3D), and the results of the CCK-8 assays indicated that the proliferative ability of ccRCC cells was obviously decreased by TLN2 overexpression when compared with the control group (Figure 3E,3F).
Effect of overexpression of TLN2 on ccRCC cell growth and metastasis in vivo
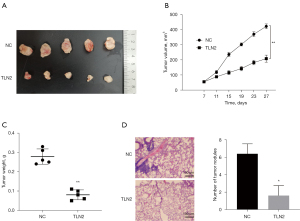
According to data described above, TLN2 overexpression resulted in obviously impaired proliferative ability of ccRCC cells, so we further investigated whether TLN2 overexpression affected ccRCC growth in vivo. We first established a mouse xenograft model by subcutaneously injecting ccRCC cells with and without TLN2 overexpression into nude mice. Our results demonstrated that tumor growth in the TLN2 overexpression group was markedly impaired compared with the control group (Figure 4A,4B). Moreover, TLN2 overexpression led to a notable reduction in tumor weight compared with the control group (Figure 4C). We further explored whether TLN2 inhibited ccRCC cell metastasis in vivo. Results showed that overexpression of TLN2 resulted in smaller nodules and decreased metastatic foci in the lung (Figure 4D). Taken together, these results showed that TLN2 overexpression resulted in notable inhibition of cell proliferation and lung metastasis in vivo.
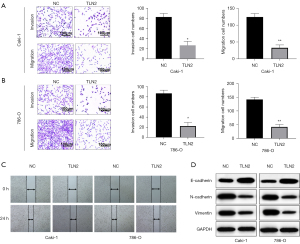
Effect of TLN2 upregulation on ccRCC cell migration and invasion
As metastasis also plays a vital role in tumor progression, we further examined the effect of TLN2 on ccRCC cell migration and invasion. Transwell assays revealed that TLN2 upregulation obviously enhanced the invasive and migration capacities of Caki-1 cells (Figure 5A). Similarly, these capacities of 786-O cells were also increased after TLN2 overexpression (Figure 5B). Moreover, as demonstrated by the wound healing assays, upregulation of TLN2 impaired the migration of Caki-1 and 786-O cells (Figure 5C). Epithelial-mesenchymal transition (EMT) is the process that epithelial cells lose the apical-basal polarity and cell-cell adhesion, and transit to invasive mesenchymal cells. EMT has received increasing attention for its role in cancer. To further confirm the function of TLN2 in ccRCC metastasis, the expressions of EMT markers were evaluated by western blot. We found that TLN2 overexpression increased E-cadherin expression, but reduced the expression of N-cadherin, and vimentin (Figure 5D).
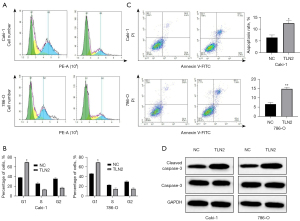
Effect of TLN2 overexpression on cell cycle progression and apoptosis of ccRCC cells
To elucidate the mechanisms underlying TLN2 suppression of ccRCC cell proliferation, we explored the effects of TLN2 on the ccRCC cell cycle. According to the results of flow cytometry, the percentage of Caki-1-TLN2 cells in the G0/G1 phase was obviously enhanced (Figure 6A). The results also revealed that TLN2 upregulation could induce cell cycle arrest in the G0/G1 phase in 786-O cells (Figure 6B). Moreover, apoptosis assays were performed to verify whether cell apoptosis regulated by TLN2 was a result of cell cycle arrest. There was significantly increased apoptosis of Caki-1 and 786-O cells after TLN2 transfection (Figure 6C). Furthermore, the levels of apoptosis-related genes were examined by western blot. We found an obvious increase in cleaved caspase-3 in TLN2-overexpressed Caki-1 and 786-O cells, while the levels of total caspase-3 showed no significant changes (Figure 6D). In general, the findings revealed that TLN2 could promote ccRCC cell apoptosis and induce cell cycle arrest in the G0/G1 phase.
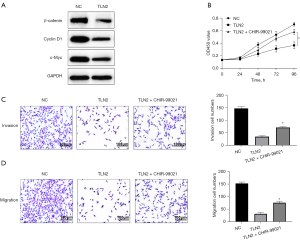
Inhibition of ccRCC tumorigenesis largely by TLN2 regulation of Wnt/β‑catenin signaling
We further investigated the potential mechanisms by which TLN2 inhibited ccRCC tumorigenesis. Firstly, the association between TLN2 and Wnt/β‑catenin signaling was analyzed by western blot analysis. We found that TLN2 overexpression notably suppressed the expressions of activated β-catenin, cyclin D1 and c-Myc in Caki-1 cells (Figure 7A). Furthermore, to confirm that the inhibitory effect on ccRCC tumorigenesis mediated by TLN2 was regulated by a Wnt/β‑catenin signaling-dependent pathway, TLN2-overexpressed cells were treated with CHIR-99021, which activates Wnt/β-catenin signaling. The results of the CCK-8 assays showed that treatment with CHIR-99021 could restore cell proliferation that was inhibited by TLN2 overexpression (Figure 7B). Similarly, TLN2 overexpression failed to inhibit Caki-1 cell invasion and migration after Wnt/β-catenin signaling activation by CHIR-99021 (Figure 7C,7D).
Discussion
ccRCC is the most common histological type of kidney cancer (20). However, the unclear mechanism of the occurrence and development of ccRCC also poses great difficulties for the development of new drugs for ccRCC treatment (21). In recent years, more and more proteins related to the occurrence and development of ccRCC have been discovered thanks to progress in molecular biology (22). The discovery of protein markers not only helps the prognostic assessment of ccRCC patients, but also helps the clinical diagnosis of ccRCC, prediction of postoperative local recurrence or distant metastasis, and is also expected to be a target for the development of molecularly targeted drugs for clinical treatment (23,24). Therefore, the search for new genes associated with ccRCC will help explain its pathogenesis and improve its diagnosis and treatment.
Infiltrative growth and metastasis of tumor cells is a major obstacle to cancer treatment and a major contributor to cancer-related death (25). The molecular regulatory mechanisms of malignant metastasis are poorly understood, resulting in most malignant tumors being difficult to treat curatively. The ECM located around tumor cells is a fundamental component of the tumor microenvironment and contains a variety of growth factors and cytokines (26). Tumor cells adhere to the ECM and metastasize through degradation of the ECM, while also providing essential growth factors for tumor growth (27). Talin is a key molecule in the ECM, integrins and cytoskeleton, and a change from the inactive to the activated state of integrins is mainly due to intracellular Talin (28). In primary tumors and their metastases, integrins bind to collagen deposited during the pro-fibroproliferative response and accelerate tumor cell proliferation, survival, chemoresistance and metastasis (29). Talin binds directly to the β-integrin cytoplasmic tail and then directly connects integral proteins to the cytoskeleton through its actin-binding domain, which plays a crucial role in cancer growth and metastasis (30). Little is known about the role of TLN2 in the mechanism of tumor invasion and metastasis.
In present study, the role of TLN2 in ccRCC progression was investigated. We found that TLN2 was downregulated in ccRCC tissues, which indicated a poorer prognosis and correlated with malignant clinicopathological characteristics. Moreover, our results also suggested that TLN2 upregulation in ccRCC cells suppressed cell viability and caused cell cycle arrest in the G1 phase. Additionally, the results of in vivo and in vitro metastasis assays confirmed that TLN2 overexpression inhibited ccRCC metastasis. Investigations of the mechanism of TLN2 inhibition of ccRCC metastasis showed that overexpression of TLN2 inhibited EMT. Previous studies have shown that the Wnt/β‑catenin pathway is implicated in the tumorigenesis of different tumors, so we hypothesized that there may be an association between TLN2 and Wnt/β‑catenin in ccRCC. Our results did indicate that TLN2 overexpression decreased the expressions of activated β-catenin, cyclin D1 and c-Myc. Furthermore, to confirm that the inhibitory function in ccRCC tumorigenesis mediated by TLN2 was regulated by Wnt/βcatenin signaling-dependent pathway, TLN2-overexpressed cells were treated with CHIR-99021, which activates Wnt/β-catenin signaling. We found that the Wnt signaling promoter blocked ccRCC proliferation, migration and invasion with concurrent overexpression of TLN2.TLN2 and TLN1 encode proteins with 74% sequence identity (31). Expression levels of both TLN2 and TLN1 were found to be significantly altered in many cancers. TLN1 is a cytoskeleton protein that participates in migration and plays a role in tumor formation, migration, and metastasis in different cancer types. Studies by Fang et al. showed that TLN2 and TLN1 exhibited lower expressions in liver cancer cells than in normal liver cell line and TLN1 overexpression could inhibited the invasion and migration abilities of liver cancer cells (32). A study also discovered that TLN-2 expression levels were also decreased in stable TLN-1 knockdown cells. Taken together, these findings suggest that TLN2 and TLN1 might regulate ccRCC invasion and migration through complex mechanisms that are not completely understood.
In conclusion, our findings demonstrated that low levels of TLN2 notably correlated with shorter survival rates and tumor aggressiveness in patients with ccRCC. Furthermore, our study provided evidence that TLN2 correlated with ccRCC proliferation, migration and invasion via regulation of the Wnt/β-catenin pathway. Our results suggested that TLN2 might be a novel therapeutic target for patients with ccRCC. Nevertheless, more in-depth research on the underlying mechanisms of TLN2 in ccRCC should be carried out in future studies; for example, the effects of EMT, etc.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at https://tau.amegroups.com/article/view/10.21037/tau-21-914/rc
Data Sharing Statement: Available at https://tau.amegroups.com/article/view/10.21037/tau-21-914/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tau.amegroups.com/article/view/10.21037/tau-21-914/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by ethics committee of Quanzhou First Hospital Affiliated to Fujian Medical University (No.: QZFH.FJMU.2018.175) and informed consent was taken from all the patients. All animal experiments were performed under a project license (No.: QZFH.FJMU.2018.a261) granted by the Animal Care and Use Committee of Quanzhou First Hospital Affiliated to Fujian Medical University, in compliance with institutional and national regulations for the care and use of animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Fuchs HE, et al. Cancer Statistics, 2021. CA Cancer J Clin 2021;71:7-33. [Crossref] [PubMed]
- Sun M, De Velasco G, Brastianos PK, et al. The Development of Brain Metastases in Patients with Renal Cell Carcinoma: Epidemiologic Trends, Survival, and Clinical Risk Factors Using a Population-based Cohort. Eur Urol Focus 2019;5:474-81. [Crossref] [PubMed]
- Yang H, Xiong X, Li H. Development and Interpretation of a Genomic Instability Derived lncRNAs Based Risk Signature as a Predictor of Prognosis for Clear Cell Renal Cell Carcinoma Patients. Front Oncol 2021;11:678253. [Crossref] [PubMed]
- Ambavane A, Yang S, Atkins MB, et al. Clinical and economic outcomes of treatment sequences for intermediate- to poor-risk advanced renal cell carcinoma. Immunotherapy 2020;12:37-51. [Crossref] [PubMed]
- Wei Q, He H, Lv L, et al. The promising role of radiotherapy in the treatment of advanced or metastatic renal cell carcinoma: a narrative review. Transl Androl Urol 2020;9:2821-30. [Crossref] [PubMed]
- Graham J, Dudani S, Heng DYC. Prognostication in Kidney Cancer: Recent Advances and Future Directions. J Clin Oncol 2018; Epub ahead of print. [Crossref] [PubMed]
- Yakirevich E, Pavlick DC, Perrino CM, et al. NF2 Tumor Suppressor Gene Inactivation in Advanced Papillary Renal Cell Carcinoma. Am J Surg Pathol 2021;45:716-8. [Crossref] [PubMed]
- Billon E, Chanez B, Rochigneux P, et al. Soluble BTN2A1 Is a Potential Prognosis Biomarker in Pre-Treated Advanced Renal Cell Carcinoma. Front Immunol 2021;12:670827. [Crossref] [PubMed]
- Zhang H, Wei P, Lv W, et al. MELK is Upregulated in Advanced Clear Cell Renal Cell Carcinoma and Promotes Disease Progression by Phosphorylating PRAS40. Cell Transplant 2019;28:37S-50S. [Crossref] [PubMed]
- Kondo K, Yao M. Characterization of Clear Cell Renal Cell Carcinoma with a Wild-type von Hippel-Lindau Tumor Suppressor Gene: Limitation of Survival Expectations Using Single-gene Alteration. Eur Urol Focus 2016;1:291-2. [Crossref] [PubMed]
- Wiele AJ, Bathala TK, Hahn AW, et al. Lenvatinib with or Without Everolimus in Patients with Metastatic Renal Cell Carcinoma After Immune Checkpoint Inhibitors and Vascular Endothelial Growth Factor Receptor-Tyrosine Kinase Inhibitor Therapies. Oncologist 2021;26:476-82. [Crossref] [PubMed]
- Ghosh N, Hossain U, Mandal A, Sil PC. The Wnt signaling pathway: a potential therapeutic target against cancer. Ann N Y Acad Sci 2019;1443:54-74. [Crossref] [PubMed]
- Wang Q, Xu J, Xiong Z, et al. CENPA promotes clear cell renal cell carcinoma progression and metastasis via Wnt/beta-catenin signaling pathway. J Transl Med 2021;19:417. [Crossref] [PubMed]
- Zhu L, Plow EF, Qin J. Initiation of focal adhesion assembly by talin and kindlin: A dynamic view. Protein Sci 2021;30:531-42. [Crossref] [PubMed]
- Kerstein PC, Patel KM, Gomez TM. Calpain-Mediated Proteolysis of Talin and FAK Regulates Adhesion Dynamics Necessary for Axon Guidance. J Neurosci 2017;37:1568-80. [Crossref] [PubMed]
- Goult BT, Xu XP, Gingras AR, et al. Structural studies on full-length talin1 reveal a compact auto-inhibited dimer: implications for talin activation. J Struct Biol 2013;184:21-32. [Crossref] [PubMed]
- Kumar A, Ouyang M, Van den Dries K, et al. Talin tension sensor reveals novel features of focal adhesion force transmission and mechanosensitivity. J Cell Biol 2016;213:371-83. [Crossref] [PubMed]
- Ji L, Jiang F, Cui X, et al. Talin1 knockdown prohibits the proliferation and migration of colorectal cancer cells via the EMT signaling pathway. Oncol Lett 2019;18:5408-16. [Crossref] [PubMed]
- Fang KP, Dai W, Ren YH, et al. Both Talin-1 and Talin-2 correlate with malignancy potential of the human hepatocellular carcinoma MHCC-97 L cell. BMC Cancer 2016;16:45. [Crossref] [PubMed]
- Internò V, De Santis P, Stucci LS, et al. Prognostic Factors and Current Treatment Strategies for Renal Cell Carcinoma Metastatic to the Brain: An Overview. Cancers (Basel) 2021;13:2114. [Crossref] [PubMed]
- Gkolfinopoulos S, Psyrri A, Bamias A. Clear-cell renal cell carcinoma - A comprehensive review of agents used in the contemporary management of advanced/metastatic disease. Oncol Rev 2021;15:530. [Crossref] [PubMed]
- Chu G, Xu T, Zhu G, et al. Identification of a Novel Protein-Based Signature to Improve Prognosis Prediction in Renal Clear Cell Carcinoma. Front Mol Biosci 2021;8:623120. [Crossref] [PubMed]
- Fan X, Liu B, Wang Z, et al. TACC3 is a prognostic biomarker for kidney renal clear cell carcinoma and correlates with immune cell infiltration and T cell exhaustion. Aging (Albany NY) 2021;13:8541-62. [Crossref] [PubMed]
- Li D, Tang L, Liu B, et al. RIPK2 is an unfavorable prognosis marker and a potential therapeutic target in human kidney renal clear cell carcinoma. Aging (Albany NY) 2021;13:10450-67. [Crossref] [PubMed]
- Zou D, Cui D. Advances in isolation and detection of circulating tumor cells based on microfluidics. Cancer Biol Med 2018;15:335-53. [Crossref] [PubMed]
- Poltavets V, Kochetkova M, Pitson SM, et al. The Role of the Extracellular Matrix and Its Molecular and Cellular Regulators in Cancer Cell Plasticity. Front Oncol 2018;8:431. [Crossref] [PubMed]
- Paolillo M, Schinelli S. Extracellular Matrix Alterations in Metastatic Processes. Int J Mol Sci 2019;20:4947. [Crossref] [PubMed]
- Haining AW, Lieberthal TJ, Del Río Hernández A. Talin: a mechanosensitive molecule in health and disease. FASEB J 2016;30:2073-85. [Crossref] [PubMed]
- Alday-Parejo B, Stupp R, Rüegg C. Are Integrins Still Practicable Targets for Anti-Cancer Therapy? Cancers (Basel) 2019;11:978. [Crossref] [PubMed]
- Sun Z, Costell M, Fässler R. Integrin activation by talin, kindlin and mechanical forces. Nat Cell Biol 2019;21:25-31. [Crossref] [PubMed]
- Praekelt U, Kopp PM, Rehm K, et al. New isoform-specific monoclonal antibodies reveal different sub-cellular localisations for talin1 and talin2. Eur J Cell Biol 2012;91:180-91. [Crossref] [PubMed]
- Fang KP, Zhang JL, Ren YH, et al. Talin-1 correlates with reduced invasion and migration in human hepatocellular carcinoma cells. Asian Pac J Cancer Prev 2014;15:2655-61. [Crossref] [PubMed]


