
Testis-enriched Asb12 is not required for spermatogenesis and fertility in mice
Introduction
To ensure the faithful transmission of genetic information to offspring, germ cells undergo dramatic dynamic remodeling throughout spermatogenesis. This process involves the self-renewal of spermatogonial stem cells (SSCs), meiosis, and spermatid maturation (1). Spermatogenesis is an intricate process and coordinated carefully by an elaborate network of genes (2,3). A total of 2,300 genes have been identified as testis-enriched genes in mice (4). Clarifying the role of these genes during spermatogenesis can reveal the molecular mechanisms underlying spermatogenesis, and assist in understanding the etiology of infertility and contraception in males. However, the function of many testis-enriched genes is not known.
Previously, using a gene-knockout (KO) approach mediated by the clustered regularly interspaced short palindromic repeats/CRISPR-associated protein 9 (CRISPR/Cas9) system, we generated mice with deleted testis-enriched genes, and our data demonstrated some of them to be crucial for spermatogenesis. For example: T-complex-associated-testis-expressed 1 (TCTE1) is an important axonemal protein that is required for sperm motility (5); F-box protein 47 (FBXO47) is localized specifically in spermatocytes and directs integration between the telomere and inner nuclear envelope during meiotic progression (6); MRN complex interacting protein (MRNIP) is a MRN-interacting protein essential for chromosome synapsis and meiotic homologous recombination (7). On the other hand, we have also shown that several genes [Retinoic acid induced 14 (Rai14), testis expressed 33 (Tex33), Fibronectin type 3 and ankyrin repeat domains 1 (Fank1), and Eukaryotic elongation factor-2 kinase (Eef2k)], previously postulated as crucial factors for spermatogenesis, are dispensable for spermatogenesis and male fertility (8-11).
The ankyrin repeat and suppressor of cytokine signaling (SOCS) box (Asb) protein family contains 18 members in humans and mice. All of them have N-terminal ankyrin (ANK) repeats and a C-terminal SOCS box (12,13). ANK is required for substrate recognition, whereas the SOCS domain interacts with elongin B/C to mediate the ubiquitination and degradation of substrates (14). Recent evidence has revealed a definitive role for the Asb family during spermatogenesis. For example, Asb3, Asb4, Asb8, Asb9, and Asb17 have been reported to be expressed abundantly in mouse testes (15-19); Asb1-KO mice exhibit hypospermatogenesis with an apparent loss of germ cells in seminiferous tubules (15); deficiency of Asb17 in mice can prevent spermatogonial apoptosis through regulation of polyubiquitination of the anti-apoptotic proteins BCL2-like 2 (BCL2L2) and MCL1 apoptosis regulator, BCL2 family member (MCL1) (20). Apart from the roles of the Asb family during spermatogenesis, recent studies have indicated Asb members to be involved in development of the central nervous system (21), myogenesis (22), as well as tumorigenesis (23).
In the present study, we identified Asb12 to be a novel testis-enriched gene in mice. We wished to uncover the role of Asb12 in murine spermatogenesis using an Asb12-KO model. We present the following article in accordance with the ARRIVE reporting checklist (available at https://tau.amegroups.com/article/view/10.21037/tau-21-900/rc).
Methods
Ethical approval of the study protocol
This study was carried out in accordance with the Guide for the Care and Use of Laboratory Animals published by the US National Institutes of Health (Bethesda, MD, USA). Animal experiments were approved (license number: 2004020) by the Animal Ethical and Welfare Committee of Nanjing Medical University (Nanjing, China).
Mice
Asb12-KO mice were generated using the CRISPR/Cas9 method according to a protocol reported previously by our research team (8,10). Single guide (sg)RNA was designed to target exon 4 of Asb12: 5'-GTAGATGCTACCACTAGGACAGG-3'. The target sequence was 5'-CCTGTCCTAGTGGTAGCATCTAC-3'. Generation of Cas9 mRNA was done as described previously (24,25). CD-1 mouse zygotes were co-microinjected with sgRNA and Cas9 mRNA. Founders were backcrossed with CD-1 mice to produce heterozygous mice. Pure-background CD-1 Asb12-KO and Asb12-wild-type (WT) mice were used for subsequent experiments. The genotype was identified by polymerase chain reaction (PCR) and subsequent Sanger sequencing using the forward primer 5'-AAGGAGGAGGAGGACACT-3' and reverse primer 5'-AGATGGAAGGTAGATGTTAGC-3'. Experimental animals were maintained under a specific pathogen-free condition at the Animal Center of Nanjing Medical University.
Fertility test
Three paired male (aged 8 weeks) Asb12 (WT and KO) mice were caged, respectively, with WT females (aged 8 weeks) at a ratio of 1:2 for 2 months. The litter size and the number of pups born were recorded.
RNA extraction and real-time reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA from testicular tissues was extracted using TRIzol™ Reagent (Invitrogen, Carlsbad, CA, USA) following manufacturer instructions. Then, RNA was reverse-transcribed into complementary (c)DNA and processed using a real-time PCR system (Applied Biosystems, Foster City, CA, USA) for quantification. 18S rRNA served as an internal control. The primers for each gene are summarized in Table S1.
Histology
Testes or epididymides from Asb12-WT and Asb12-KO mice of age 8–12 weeks were collected and fixed in modified Davidson’s fluid (MDF) for 48 h. Then, tissues were dehydrated in a graded series of ethanol solutions, embedded in paraffin, and sectioned at 4-μm thickness. These sections were rehydrated and stained with hematoxylin and eosin (H&E) or periodic acid Schiff (PAS). Next, we undertook computer-assisted sperm analysis (CASA). Briefly, sperm from Asb12-WT and Asb12-KO mice aged 8–12 weeks were collected from the cauda epididymis and suspended in culture medium. Semen quality was assayed using the Ceros™ II Sperm Analysis System (Hamilton Thorne, Beverly, MA, USA). For ultrastructural analysis, sperm samples from Asb12-WT and Asb12-KO mice aged 8–12 weeks were fixed in 2% (v/v) glutaraldehyde overnight and immersed in 2% (w/v) OsO4 for 2 h. Next, samples were embedded in araldite and sectioned at 80-nm thickness. Images were captured under a transmission electron microscope (JEM-1410; JEOL, Tokyo, Japan).
Immunofluorescence
Sections were deparaffinized and rehydrated, followed by antigen retrieval in sodium citrate buffer as described previously (26,27). After blockade with 1% (w/v) bovine serum albumin for 2 h, samples were reacted with the indicated primary antibodies (Table S2) overnight at 4 °C. Nonimmune immunoglobulin (Ig)G antibodies were used for the negative controls (Figure S1). Subsequently, slides were washed thrice with phosphate-buffered saline (PBS), and then probed with Alexa-Fluor secondary antibodies (Thermo Scientific, Waltham, USA) for 1 h at 37 °C. Finally, sections were counterstained with 4′,6-diamidino-2-phenylindole (Beyotime Institute of Biotechnology, Nantong, China) and subjected to fluorescence analyses under a LSM800 confocal microscope (Zeiss, Oberkochen, Germany).
Terminal deoxynucleotidyl transferase-dUTP nick-end labeling (TUNEL) assay
Apoptosis was detected using a TUNEL kit (Vazyme, Nanjing, China) following manufacturer instructions. Briefly, paraffin sections were incubated with proteinase K for 15 min before being allowed to react with TUNEL labeling mix buffer for 1 h at 37 °C. Apoptotic cells were counted under a confocal laser scanning microscope (LSM800; Zeiss).
Chromosome spread
Preparation of chromosome spreads was conducted as described previously (28). Briefly, testicular tissues from Asb12-WT and Asb12-KO mice aged 8–12 weeks were digested by collagenase and trypsin for 15 min at 37 °C before immersion in a hypotonic extraction buffer. Then, cell pellets were suspended in sucrose (100 mM) and spread on slides covered with fixative solution. After fixation, samples were washed with PBS and subjected to immunostaining with the indicated primary antibodies (Table S2).
Statistical analyses
Statistical analyses were conducted using Prism 8.0 (GraphPad, La Jolla, CA, USA). Data are the mean ± SD from at least three independent experiments. Biological replicates are indicated in the corresponding figure legends. The unpaired Student’s t-test or chi-square test were used to calculate the differences between Asb12-WT and Asb12-KO mice.
Results
Generation of Asb12-KO mice
ASB12 belongs to the ASB protein family, in which all members have the classic ANK and SOCS domains. Bioinformatic analysis revealed ASB12 to be an evolutionarily conserved protein with conservation of the ANK and SOCS domains among species (Figure S2A). RT-qPCR revealed Asb12 to be expressed abundantly in the mouse testis (Figure S2B). These findings suggested that Asb12 might have a conserved role during spermatogenesis.
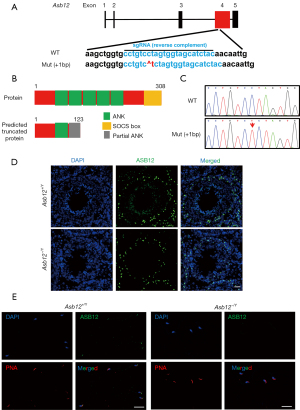
To explore the function of Asb12 in vivo, Asb12 mutant mice were generated by CRISPR/Cas9-mediated gene editing which, ultimately, caused a frameshift mutation in exon 4 of Asb12 on the X chromosome by a 1-bp insertion (Figure 1A). This mutation is predicted to result in a truncated protein of 123 amino acids, causing a partial loss of the ANK domain and complete loss of SOCS domains (Figure 1B). The mutation was confirmed by PCR and Sanger sequencing (Figure 1C). Furthermore, using immunostaining of the ASB12 protein in mouse testicular tissue and mature sperm, we observed an obvious location of ASB12 in the acrosome of Asb12-WT (+/Y) mice. In contrast, there was no detectable signal in the acrosome of Asb12 mutant mice (Figure 1D,1E), suggesting an absence of the ASB12 protein in the testes of Asb12 mutant mice. The fluorescence signal in the basal compartment of the seminiferous epithelium was probably non-specific because it was also present in the testes of Asb12 mutant mice (Figure 1D). Thus, we first successfully constructed an Asb12-KO (−/Y) mouse strain. The specific location of ASB12 in mouse sperm suggested that ASB12 may be associated with acrosome maturation during spermatogenesis.
Normal fertility and semen quality in Asb12-KO mice
Asb12-KO mice developed and survived normally. Asb12-KO males were fertile and led to the production of similar litter sizes to that obtained with Asb12-WT mice (Figure 2A). The testes of Asb12-WT and Asb12-KO mice were comparable in size and weight (Figure 2B,2C). CASA revealed Asb12-KO mice to have a normal concentration, motility, and progressive ratio of sperm compared with those of Asb12-WT mice (Figure 2D-2F). H&E staining of sperm revealed Asb12-KO mice to exhibit normal sperm morphology compared with that of Asb12-WT mice (Figure 2G,2H). Due to the specific location of ASB12 in the acrosome of sperm, we undertook ultrastructural analyses of sperm in Asb12-WT and Asb12-KO mice. Ultrastructurally, Asb12-KO sperm displayed normal morphology with a well-formed acrosome, condensed head, and typical “9 + 2” axoneme (Figure 2I). Moreover, no significant change of progesterone-induced acrosome reaction between WT and KO was observed (Figure S3). We therefore concluded that Asb12 was not required for fertility and semen quality in mice.

Normal spermatogenesis in Asb12-KO mice
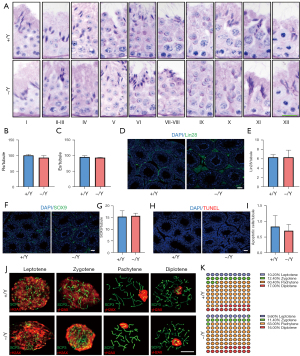
Testicular morphology was examined by H&E staining. Asb12-WT and Asb12-KO mice exhibited intact seminiferous tubules with all stages of spermatogenic cells (Figure 3A). Moreover, there were no obvious histological changes of the epididymis between Asb12-WT and Asb12-KO mice (Figure 3B,3C). These data suggested that Asb12 deficiency did not have an effect on spermatogenesis.

Spermatogenesis has three main processes: SSC (spermatogonial stem cell) proliferation, meiosis, and spermiogenesis. To determine precisely the process of spermatogenesis in the testes of Asb12-KO mice, we first undertook PAS staining to analyze the dynamics of acrosome development and nucleus morphology during spermiogenesis. There was no measurable morphological difference in spermatids between Asb12-WT and Asb12-KO mice during spermiogenesis (Figure 4A). In addition, the number of round spermatids and elongating/elongated spermatids was comparable between Asb12-WT and Asb12-KO mice (Figure 4B,4C). Moreover, Lin28, SOX9, and TUNEL signals were used to quantify the number of SSCs, Sertoli cells, and apoptotic cells in testes, respectively: the number of these cell types did not differ between Asb12-WT and Asb12-KO mice (Figure 4D-4I). Finally, the chromosome-spread method was conducted to evaluate meiotic progression. Under co-immunostaining of SCP3 (a marker of the lateral element of the synaptonemal complex) and γH2AX (a marker for DNA double-strand breaks), the subdivided stages of meiotic prophase (leptotene, zygotene, pachytene, and diplotene) could be distinguished readily according to the dynamic distribution of SCP3 and γH2AX (6,7,28). However, marked morphological alteration of SCP3 and γH2AX was not observed in Asb12-KO spermatocytes when compared with that in the WT group (Figure 4J,4K). Taken together, these results indicated that Asb12 was dispensable for mouse spermatogenesis.
Functional compensation from paralogs in the testes of Asb12-KO mice
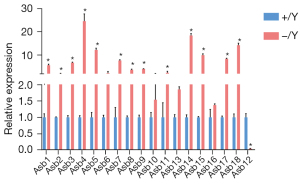
We undertook real-time RT-qPCR to assess the relative transcription levels of the 18 Asb genes in the testes of Asb12-WT and Asb12-KO mice. Interestingly, expression of Asb-1,2, 3, 4, 5, 7, 8, 9, 11, 14, 15, 17, and 18 was increased significantly in the testes of Asb12-KO mice compared with that in the testes of Asb12-WT mice (Figure 5). The relative transcription level of Asb12 was reduced markedly in the testes of Asb12-KO mice (Figure 5), which suggested that Asb12 mutation could cause a decay of the encoded mRNA. This finding strongly suggests that other members of the Asb family may compensate for Asb12 deficiency.
Discussion
A gene microarray study by Schultz et al. uncovered >2,300 testis-enriched genes in the mouse genome. Using the gene-KO approach in mice, we and other research teams have revealed many testis-enriched genes that are required for spermatogenesis and male fertility (5-7,29-31). Nevertheless, several testis-enriched genes that had been postulated previously to be essential factors for spermatogenesis have been shown to be unnecessary for spermatogenesis in KO animal models (32). Although gene-KO mice with no obvious reproductive phenotypes have rarely been reported, the genetic and phenotypic information must be disseminated to the scientific community to prevent redundant efforts by other research teams. In 2017, Castaneda et al. shared information on 54 testis-enriched genes that are not essential for spermatogenesis or fertility in mice via a genome-engineering approach. Thereafter, several large-scale screenings of non-essential testis-enriched genes were published (33,34).
Our research team has been using the CRISPR/Cas9 method to produce mice that delete testis-enriched genes. Here, we investigated the effects of the loss of a testis-enriched gene, Asb12, on spermatogenesis and fertility in mice using CRISPR/Cas9-mediated gene editing. We discovered that, despite the evolutionary conservation of Asb12 in vertebrates, it is not essential for spermatogenesis and male fertility. Although such functional redundancy renders Asb12 irrelevant as a target for contraception, the restricted expression of ASB12 in the sperm acrosome may enable it to serve as a potential biomarker for non-obstructive azoospermia.
A reasonable explanation for Asb12-KO mice displaying normal spermatogenesis and fertility may be functional redundancy. Genetic compensation upon gene KO is a common phenomenon among species. Paralogous genes with overlapping functions may compensate for the loss of one gene (35,36). For instance, KO of Ubqln3 in mouse testes induces significant upregulation of expression of Ubqln1, Ubqln2, and Ubqln4, whereas KO of UbqlnL provokes a marked increase in expression of Ubqln1 and Ubqln4 in testes (37,38). Likewise, in our study, a loss of Asb12 in mouse testes elicited a dramatic increase in expression of several Asb members. To overcome functional redundancy, generation of double-KO (or even triple-KO) models is needed to eliminate the effects of functional compensation from paralogs.
Conclusions
In this study, we generated a murine Asb12-KO model. Although Asb12 was shown to be a testis-enriched gene in mice, Asb12 itself had no obvious effects on spermatogenesis and male fertility in mice. The phenotypic information of Asb12-KO mice we have provided will assist other researchers in prioritizing their resources to focus on genes that are crucial for male fertility.
Acknowledgments
Funding: This work was supported by the National Natural Science Foundation of China (No. 81901532 to BZ), Natural Science Foundation of Jiangsu Province (No. BK20190188 to BZ, No. BK20190081 to ML), Open Fund of State Key Laboratory of Reproductive Medicine of Nanjing Medical University (No. SKLRM-K202010 to BZ), and Gusu Health Talent Program of Suzhou (No. GSWS2020068 to BZ).
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at https://tau.amegroups.com/article/view/10.21037/tau-21-900/rc
Data Sharing Statement: Available at https://tau.amegroups.com/article/view/10.21037/tau-21-900/dss
Peer Review File: Available at https://tau.amegroups.com/article/view/10.21037/tau-21-900/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tau.amegroups.com/article/view/10.21037/tau-21-900/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was carried out in accordance with the
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Wu S, Yan M, Ge R, et al. Crosstalk between Sertoli and Germ Cells in Male Fertility. Trends Mol Med 2020;26:215-31. [Crossref] [PubMed]
- Wen L, Tang F. Human Germline Cell Development: from the Perspective of Single-Cell Sequencing. Mol Cell 2019;76:320-8. [Crossref] [PubMed]
- Tan K, Wilkinson MF. A single-cell view of spermatogonial stem cells. Curr Opin Cell Biol 2020;67:71-8. [Crossref] [PubMed]
- Schultz N, Hamra FK, Garbers DL. A multitude of genes expressed solely in meiotic or postmeiotic spermatogenic cells offers a myriad of contraceptive targets. Proc Natl Acad Sci U S A 2003;100:12201-6. [Crossref] [PubMed]
- Castaneda JM, Hua R, Miyata H, et al. TCTE1 is a conserved component of the dynein regulatory complex and is required for motility and metabolism in mouse spermatozoa. Proc Natl Acad Sci U S A 2017;114:E5370-8. [Crossref] [PubMed]
- Hua R, Wei H, Liu C, et al. FBXO47 regulates telomere-inner nuclear envelope integration by stabilizing TRF2 during meiosis. Nucleic Acids Res 2019;47:11755-70. [Crossref] [PubMed]
- Lin M, Lv J, Zhao D, et al. MRNIP is essential for meiotic progression and spermatogenesis in mice. Biochem Biophys Res Commun 2021;550:127-33. [Crossref] [PubMed]
- Wu Y, Wang T, Zhao Z, et al. Retinoic Acid Induced Protein 14 (Rai14) is dispensable for mouse spermatogenesis. PeerJ 2021;9:e10847. [Crossref] [PubMed]
- Zhu Z, Zhang X, Zeng W, et al. Spermatogenesis is normal in Tex33 knockout mice. PeerJ 2020;8:e9629. [Crossref] [PubMed]
- Zhang J, Zhang X, Zhang Y, et al. Normal spermatogenesis in Fank1 (fibronectin type 3 and ankyrin repeat domains 1) mutant mice. PeerJ 2019;7:e6827. [Crossref] [PubMed]
- Feng T, Zhou S, Shi X, et al. Eef2k is not required for fertility in male mice. Transl Androl Urol 2021;10:1988-99. [Crossref] [PubMed]
- Kile BT, Schulman BA, Alexander WS, et al. The SOCS box: a tale of destruction and degradation. Trends Biochem Sci 2002;27:235-41. [Crossref] [PubMed]
- Piessevaux J, Lavens D, Peelman F, et al. The many faces of the SOCS box. Cytokine Growth Factor Rev 2008;19:371-81. [Crossref] [PubMed]
- Liu P, Verhaar AP, Peppelenbosch MP. Signaling Size: Ankyrin and SOCS Box-Containing ASB E3 Ligases in Action. Trends Biochem Sci 2019;44:64-74. [Crossref] [PubMed]
- Kile BT, Metcalf D, Mifsud S, et al. Functional analysis of Asb-1 using genetic modification in mice. Mol Cell Biol 2001;21:6189-97. [Crossref] [PubMed]
- Kim SK, Rhim SY, Lee MR, et al. Stage-specific expression of ankyrin and SOCS box protein-4 (Asb-4) during spermatogenesis. Mol Cells 2008;25:317-21. [PubMed]
- Lee MR, Kim SK, Kim JS, et al. Expression of murine Asb-9 during mouse spermatogenesis. Mol Cells 2008;26:621-4. [PubMed]
- Guo JH, Saiyin H, Wei YH, et al. Expression of testis specific ankyrin repeat and SOCS box-containing 17 gene. Arch Androl 2004;50:155-61. [Crossref] [PubMed]
- Kim KS, Kim MS, Kim SK, et al. Murine Asb-17 expression during mouse testis development and spermatogenesis. Zygote 2004;12:151-6. [Crossref] [PubMed]
- Yang G, Wan P, Xiang Q, et al. E3 Ubiquitin Ligase ASB17 Promotes Apoptosis by Ubiquitylating and Degrading BCLW and MCL1. Biology (Basel) 2021;10:234. [Crossref] [PubMed]
- Diks SH, Bink RJ, van de Water S, et al. The novel gene asb11: a regulator of the size of the neural progenitor compartment. J Cell Biol 2006;174:581-92. [Crossref] [PubMed]
- Tee JM, Sartori da Silva MA, Rygiel AM, et al. asb11 is a regulator of embryonic and adult regenerative myogenesis. Stem Cells Dev 2012;21:3091-103. [Crossref] [PubMed]
- Tokuoka M, Miyoshi N, Hitora T, et al. Clinical significance of ASB9 in human colorectal cancer. Int J Oncol 2010;37:1105-11. [PubMed]
- Zhang Y, Guo R, Cui Y, et al. An essential role for PNLDC1 in piRNA 3' end trimming and male fertility in mice. Cell Res 2017;27:1392-6. [Crossref] [PubMed]
- Wang X, Xie W, Yao Y, et al. The heat shock protein family gene Hspa1l in male mice is dispensable for fertility. PeerJ 2020;8:e8702. [Crossref] [PubMed]
- Gao T, Lin M, Shao B, et al. BMI1 promotes steroidogenesis through maintaining redox homeostasis in mouse MLTC-1 and primary Leydig cells. Cell Cycle 2020;19:1884-98. [Crossref] [PubMed]
- Zhao D, Shen C, Gao T, et al. Myotubularin related protein 7 is essential for the spermatogonial stem cell homeostasis via PI3K/AKT signaling. Cell Cycle 2019;18:2800-13. [Crossref] [PubMed]
- Gao T, Lin M, Wu Y, et al. Transferrin receptor (TFRC) is essential for meiotic progression during mouse spermatogenesis. Zygote 2021;29:169-75. [Crossref] [PubMed]
- Greenbaum MP, Yan W, Wu MH, et al. TEX14 is essential for intercellular bridges and fertility in male mice. Proc Natl Acad Sci U S A 2006;103:4982-7. [Crossref] [PubMed]
- Fujihara Y, Noda T, Kobayashi K, et al. Identification of multiple male reproductive tract-specific proteins that regulate sperm migration through the oviduct in mice. Proc Natl Acad Sci U S A 2019;116:18498-506. [Crossref] [PubMed]
- Ikawa M, Wada I, Kominami K, et al. The putative chaperone calmegin is required for sperm fertility. Nature 1997;387:607-11. [Crossref] [PubMed]
- Okabe M. Mechanism of fertilization: a modern view. Exp Anim 2014;63:357-65. [Crossref] [PubMed]
- Lu Y, Oura S, Matsumura T, et al. CRISPR/Cas9-mediated genome editing reveals 30 testis-enriched genes dispensable for male fertility in mice†. Biol Reprod 2019;101:501-11. [Crossref] [PubMed]
- Park S, Shimada K, Fujihara Y, et al. CRISPR/Cas9-mediated genome-edited mice reveal 10 testis-enriched genes are dispensable for male fecundity. Biol Reprod 2020;103:195-204. [Crossref] [PubMed]
- Krakauer DC, Plotkin JB. Redundancy, antiredundancy, and the robustness of genomes. Proc Natl Acad Sci U S A 2002;99:1405-9. [Crossref] [PubMed]
- El-Brolosy MA, Stainier DYR. Genetic compensation: A phenomenon in search of mechanisms. PLoS Genet 2017;13:e1006780. [Crossref] [PubMed]
- Yuan S, Qin W, Riordan CR, et al. Ubqln3, a testis-specific gene, is dispensable for embryonic development and spermatogenesis in mice. Mol Reprod Dev 2015;82:266-7. [Crossref] [PubMed]
- Yuan S, Swiggin HM, Zheng H, et al. A testis-specific gene, Ubqlnl, is dispensable for mouse embryonic development and spermatogenesis. Mol Reprod Dev 2015;82:408-9. [Crossref] [PubMed]


