
Overexpression of F-box and WD repeat domain containing 7 prevents tumor growth of bladder cancer cells through regulating SREBP1a
Introduction
Bladder cancer is a common malignant tumor, especially in women (1), and possesses the highest incidence rate of urologic malignancy in China. It is the second most common urogenital tumors worldwide and has the ninth highest rate of cancer incidence (2,3). However, the mortality rate of bladder cancer has only decreased by 5% in the past 15 years (4). The main reason is that the efficacy of chemotherapy for advanced bladder cancer is low, and an effective target treatment drug is still not available, as the progress and drug resistance mechanism of bladder cancer have remained unclear (5). Therefore, it is of great significance to explore new molecular markers related to the occurrence and development of bladder cancer.
Hyperactivation of sterol regulatory element binding protein (SREBP) transcription factors directly upregulates lipogenic enzymes to trigger de novo lipogenesis in tumors (4). In mammals, 3 isoforms of SREBP protein have been identified: SREBP 1a, SREBP 1c, and SREBP2. Both SREBP1a and SREBP1c are produced by the SREBP1 gene, while the SREBP2 gene encodes SREBP2 (5). While SREBP1 mainly regulates the genes of fatty acid triglyceride and phosphate synthesis, SREBP2 preferentially activates cholesterol synthesis genes (5,6). Therefore, SREBP is essential to maintain the balance between protein and lipid biosynthesis and is involved in the development, migration, or poor prognosis of cancers (7,8). The imbalance of SREBP1 activates high-level de novo adipogenesis in cancer cells, which is one of the key factors driving the growth of malignant tumors (6). A new phosphorylation site ser-434 was identified in SREBPs, which regulates the degradation of SREBP1-dependent F-box and WD repeat domain containing 7 (FBXW7) (7). Interestingly, the expression of SREBP1 depends on glycogen synthase kinase-3 β (GSK-3β) activity (9,10).
A member of the F-box protein family, FBXW7 is a substrate receptor of SKP1-CUL1-F-box protein (SCF) ubiquitin ligase, which plays an important role in regulating several important cellular processes, including cell growth, division, and differentiation (8,11). It is recognized as a typical tumor suppressor because it can promote ubiquitination and degradation of a variety of cancer proteins (including cyclin E, c-myc, c-jun, Notch1, and Mcl-1), thereby inhibiting the growth and survival of tumor cells (11). Function loss of FBXW7 is often reported in cancer (8,9). Therefore, it is very important to evaluate the effect of FBXW7 in cancer treatment. In this study, we aimed to explore the effect of FBXW7 overexpression on bladder cancer cells, and to determine the regulatory effect of FBXW7 on SREBP1a in bladder cancer. In doing so, we aimed to determine novel therapeutic targets for bladder cancer. We present the following article in accordance with the ARRIVE reporting checklist (available at https://tau.amegroups.com/article/view/10.21037/tau-22-146/rc).
Methods
Cell lines
Human bladder cancer cells J28 and T24 were purchased from BeiNa Biotech and cultured in Dulbecco’s Modified Eagle Medium (DMEM; Gibco, Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS; Hyclone, Logan, UT, USA) in an incubator with 5% CO2 at 37 ℃. The experiments were divided into 3 groups: a control group (without transfection), a control vector group (NC, non-functional version of FBXW7), and a FBXW7 overexpression group (FBXW7).
Cell transfection
The FBXW7 vector was constructed in pcDNA3.1 (+) plasmid. The J28 and T24 cells were transfected with a FBXW7 vector or control vector, respectively. When the cell confluence reached 70%, the transfection was carried out. The cell culture medium was replaced with serum-free medium, and combined with opti MEM (Thermo Fisher Scientific, Waltham, MA, USA), Lipofectamine 3000 (Thermo Fisher), and small interfering RNA [siRNA; 125 µL/1 optical density (OD)]. At 4 h after transfection, the cells were refreshed with 1 mL of complete medium with 20% FBS, and the corresponding detection was carried out 48 h later.
Real-time fluorescent quantitative polymerase chain reaction
At 48 h after transfection, the cells were collected to extract total RNA using the TRIzol™ Plus RNA Purification Kit (CW0580S; CWBIO, Cambridge, MA, USA). The purity of RNA (OD260/OD280) was determined using a UV-Vis spectrophotometer (DeNovix, Wilmington, DE, USA). Complementary DNA (cDNA) was synthesized by a High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific). Thereafter, fluorescence quantitative polymerase chain reaction (qPCR) was carried out using the following reaction system: RNase free dH2O 8.2 µL, cDNA 1 µL, upstream primer 0.4 µL, downstream primer 0.4 µL, and universal SYBR qPCR Master Mix 10 µL. The reaction steps were as follows: pre-denaturation at 95 ℃ for 10 min; denaturation at 95 ℃ for 10 s; annealing at 58 ℃ for 30 s; and extension at 72 ℃ for 30 s, for a total of 40 cycles. The primer sequence was obtained from General Biosystems Co., Ltd (Anhui, China), and was as follows: FBXW7 (F: 5'-AGCTGTCCAGCCCCTTCTA-3'; R: 5'-GCACGCTTGTGATTCTCCTT-3'); SREBP1a (F: 5'-GCCTATTTGACCCACCCTAT-3'; R: 5'-TGGCACTGACTCTTCCTTGA-3'); β-actin (F: 5'-TGGCACCCAGCACAATGAA-3'; R: 5'-CTAAGTCATAGTCCGCCTAGAAGCA-3'). The relative expression levels of FBXW7 and SREBP1a were calculated by 2-∆CT method and normalized to β-actin.
Western blotting
At 48 hours after transfection, the cells were collected to isolate the proteins. After 30 min pyrolysis on ice, the homogenates were centrifuged at 12,000 rpm for 15 min. The supernatant was collected, and protein concentration was determined by bicinchoninic acid (BCA) kit. Later, the protein was denatured, and 25 µg protein was separated via 12% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) for 2 h. After that, the proteins were transferred to a nitrocellulose membrane. The rabbit anti-FBXW7 (1:1,000), rabbit anti-p-GSK3β (1:400), and mouse anti-SREBP1a (1:500) was incubated overnight at 4 ℃. The secondary antibody (1:100; catalogue nos. ab131368; Abcam, Cambridge, MA, USA) was added and co-incubated for 1–2 h at room temperature. Enhanced chemiluminescence (ECL) exposure liquid droplet (catalogue no. RPN2133; GE Healthcare Life Sciences, Chalfont, UK) was added on the membrane. Finally, the membrane was used for exposure utilizing a gel imaging system (Bio-Rad Laboratories, Inc., Hercules, CA, USA). Grey density was analyzed using Quantity One analysis software v1.4.6 (Bio-Rad Laboratories, Inc.).
Cell Counting Kit-8 (CCK-8)
CCK-8 assay was used to determine the cell proliferation as previously described (10). At 48 h after cell transfection, 10 µL CCK-8 detection reagent was added to each well and incubated at 37 ℃ for 2 h; The OD value of each well at 450 nm wavelength was detected by microplate reader to calculate the cell survival rate.
Migration assay
When the cell confluence reached more than 90%, the migration assay was carried out. A 200 µL pipette tip was used to scratch each well. The culture medium was discarded and washed with phosphate-buffered saline (PBS) 3 times. After changing to DMEM incomplete culture medium, the cells of each well were photographed. The cells were put back into the incubator, and the scratches of each well were photographed again 24 h later. The 24 h data and 0 h data were determined, and the corresponding width was calculated.
Transwell assay
The J28 and T24 cells were seeded into the upper chamber with 1-day starvation of serum. The lower chamber contained DMEM with 10% FBS. The transwell system was placed in a CO2 incubator at 37 ℃ for 24 h incubation. After that, the lower chamber was taken out and fixed with 4% paraformaldehyde (PFA) for 30 min at room temperature. The cells were stained with crystal violet (0.1%, C8470; Amresco, Solon, OH, USA) at room temperature for 5 min. At least 5 fields in each image were taken using light microscopy.
Flow cytometry
The J28 and T24 cells were seeded in 6-well plates. Approximately 3×106 cells in each group were collected after transfection. After washing with pre-cooled PBS, the cells were incubated with propidium iodide (PI) for 30 min. The cell cycles were detected using a flow cytometer [Becton, Dickinson, and Co. (BD) Biosciences, Franklin Lakes, NJ, USA] within 1 h.
In vivo tumor model
All animal experiments were approved by the Ethics Committee of the First Affiliated Hospital of Nanchang University (approval No. 20170312), in compliance with the institutional guidelines of First Affiliated Hospital of Nanchang University for the care and use of animals. A protocol was prepared before the study without registration. Male BALB/c-nu mice (4 weeks old) were purchased from Hunan SLAC Jingda Experimental Animal Co., Ltd. (Hunan, China; license No.: scxk (Xiang) 2019-0004) and housed under specific-pathogen-free conditions that were automatically maintained at a temperature of 23±2 ℃, relative humidity of 45–65%, and with a controlled 12 h light/dark cycle. A total of 19 mice implanted with the T24 cells were randomly divided into 3groups: control group (n=7), FBXW7 NC group (n=7), and FBXW7 group (n=5), in which 1×106 normal T24 cells, FBXW7 NC T24 cells, and FBXW7 overexpressed T24 cells were inoculated into the axillary subcutaneously, respectively. At 30 days after tumor cell inoculation, the tumor tissues were collected for subsequent experiments.
Hematoxylin and eosin staining
The tumor specimens were fixed in 4% PFA in PBS (pH 7.4) at 4 ℃ overnight and then embedded in paraffin for tissue sectioning. The tissues were sectioned into 5 µm-thick sections. Subsequently, the slides were stained with hematoxylin (3%) and eosin (3%) for 5 min at room temperature. The images were captured under light microscopy.
TUNEL staining
Tumor tissues were fixed in 4% PFA in PBS (pH 7.4) at 4 ℃ overnight and then embedded in paraffin for tissue sectioning. The tissues were sectioned into 5 µm-thickness sections. Then, terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) solution was added to each section and incubated for 1.5 h at 45 ℃. Thereafter, 4’,6-diamidino-2-phenylindole (DAPI) (5 µg/mL) was added to stain the nuclei for 5 min at room temperature. The images were observed under a fluorescence microscope (magnification, ×200).
Immunohistochemistry
The tumor tissues were fixed in 4% PFA in PBS (pH 7.4) at 4 ℃ overnight and then embedded in paraffin for tissue sectioning. The tissues were sectioned into 5 µm-thick sections. The sections were blocked in 5% bovine serum albumin (BSA) for 2 h at room temperature and incubated with the primary antibodies overnight at 4 ℃, namely, SREBP1α (1:200) and p-GSK3β (1:100). The sections were incubated with the secondary antibody and stained by 3,3’-diaminobenzidine (DAB) for 5–10 min.
Statistical analyses
All data were analyzed using the software SPSS 21.0 (IBM Corp., Armonk, NY, USA) and expressed as mean ± standard deviation (SD). The significant difference among groups was analyzed by one-way analysis of variance (ANOVA) and Bonferroni’s test, with P<0.05 as the significant difference.
Results
Effect of FBXW7 overexpression on migration and invasion of bladder cancer cells
The expression of FBXW7 in J82 and T24 cells were detected by reverse transcription (RT)-qPCR and western blotting. Our results showed that the messenger RNA (mRNA; Figure 1A) and protein expression (Figure 1B) levels of FBXW7 in J82 and T24 cells were significantly increased in the FBXW7 vector group compared with those in control group and control vector group, which indicated that the FBXW7 vector was efficient to enhance FBXW7 expression.
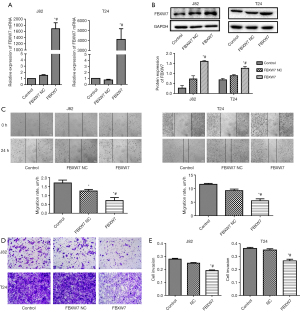
Next, we determined the effects of FBXW7 overexpression on the biological activities of the tumor cells. The results showed that overexpression of FBXW7 significantly inhibited the migration (Figure 1C), as well as the invasion of J82 and T24 cells compared with control vector group (Figure 1D), quantitative data of invasion of J82 cells and T24 cells (Figure 1E).
Effect of FBXW7 overexpression on cell cycle distribution and proliferation of bladder cancer cells
The results showed that FBXW7 could significantly affect G0/G1, S, and G2/M phases of bladder cancer J82 and T24 cells (Figure 2A), and significantly inhibit the proliferation of J82 and T24 cells compared with the control and vector group (Figure 2B). Therefore, overexpression of FBXW7 could regulate the progression of bladder cancer by affecting the cell cycle distribution and inhibiting cell proliferation.
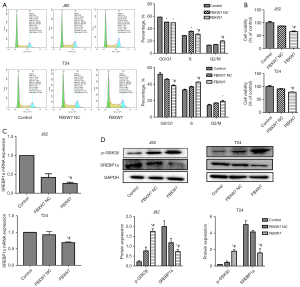
Effect of FBXW7 overexpression on expression of SREBP1a and p-GSK3β in bladder cancer cells
The expression of p-GSK3β and SREBP1a was detected. The results showed that the expression of SREBP1a in FBXW7 vector group was significantly lower than that in the control group (Figure 2C,2D), while p-GSK3β was significantly higher in FBXW7 vector group than that in the control group (Figure 2C,2D).
FBXW7 overexpression triggered apoptosis of the tumor cells
At 30 days after inoculation of different types of bladder cancer cells, the tumors were collected. Hematoxylin and eosin (HE) staining showed that tumor cells in the control and FCXW7 NC groups were diffusely distributed and pathological mitotic features were observable. In contrast, FCXW7 overexpression obviously altered the features of tumorigenesis (Figure 3A). Apoptosis increased in FBXW7 overexpression group compared with FCXW7 NC group (Figure 3B,3C).
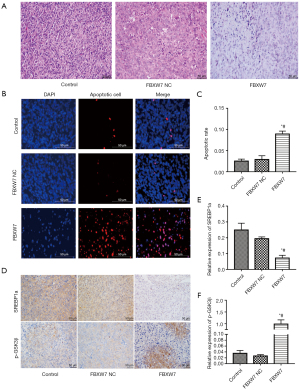
FBXW7 overexpression promoted p-GSK3β, but reduced SREBP1a expression in bladder cancer cells
Immunohistochemistry (IHC) was used to detect SREBP1a and p-GSK3β in tumor tissues. As shown in Figure 3D-3F, SREBP1a and p-GSK3β were expressed in nucleus as well as cytoplasm. Moreover, FBXW7 overexpression up-regulated p-GSK3β, but reduced SREBP1a expression in bladder cancer cells.
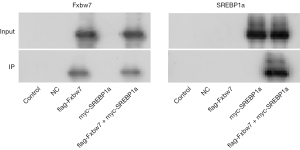
FBXW7 directly interacted with SREBP1a
Using FLAG-FBXW7 as bait protein, myc-SREBP1a protein could be detected in the product via co-immunoprecipitation experiment (Figure 4), which suggested that FBXW7 interacted with SREBP1a.
Discussion
As bladder cancer is a severe type of malignant tumor with high metastasis, it is important to determine its pathogenesis to reduce the mortality of the disease (12,13). Tumor cell migration as well as invasion is crucial for tumorigenesis (14). Importantly, the invasion of cancer cells is related to cell migration (15,16). In this study, in vitro experiments were designed to explore the effect of FBXW7 on the migration and invasion of bladder cancer cells, and to disclose its mechanism. Our results demonstrated that FBXW7 overexpression could prevent migration and invasion of bladder cancer cells. Molecular mechanisms showed that FBXW7 overexpression promoted p-GSK3 and suppressed SREBP1a expression. In vivo experiments further indicated that FBXW7 overexpression promoted tumor cell apoptosis, promoted p-GSK3, and suppressed SREBP1a expression. Moreover, a direct interaction was verified using co-immunoprecipitation. This study provides a new idea for the treatment of bladder cancer.
The FBXW gene encodes members of F-box protein family, which is the key component of skp1-cul1-f-box protein (SCF) ubiquitin ligase (17); FBXW7 targets a group of well-known oncoproteins, including c-myc, cyclin E, notch, c-jun and Mcl-1, for ubiquitination and degradation (18-20). Meanwhile, FBXW7 is involved in the regulation of many cellular processes, including cell proliferation, cell cycling, apoptosis, and differentiation (11,21,22). Previous studies have shown that FBXW7 is a tumor suppressor, because the decreased expression of FBXW7 promotes the occurrence and development of many types of cancer (23-25). Moreover, the expression level of FBXW7 is negatively correlated with the malignant progression of some tumors (25-27). It has been reported that the mutation of FBXW7 can promote the proliferation, invasion, and migration of cervical cancer cells (28), which is consistent with the results of this study. Overexpression of FBXW7 in bladder cancer cells J82 and T24 inhibits the migration, invasion, and proliferation of J82 and T24 cells, and affects the cell cycle.
As a member of SREBP transcription factor family, SREBP1a mainly regulates lipid homeostasis. It also activates high levels in de novo adipogenic tract cancer cells, which is one of the keys to driving the growth of malignant tumors (29). Phosphorylation of thr-426 and Ser-430 in SREBP1a creates a docking site for ubiquitin ligase FBXW7, leading to degradation of transcription factors (7), while Prmt5-induced methylation prevents GSK3β. The phosphorylation of SREBP1a at s430 results in its separation from FBXW7, and the degradation is avoided through ubiquitin proteasome pathway (29). In this study, overexpression of FBXW7 promoted p-GSK3β expression, while suppressing the expression of SREBP1a. Interestingly, FBXW7 has different effects on SREBP1a in J82 and T24 bladder cancer cells, and FBXW7 has a stronger effect on SREBP1a in T24 bladder cancer cells.
This study demonstrated the exact function of FBXW7 on bladder cancer cell proliferation and migration. We selected 2 bladder cancer cells, J82 and T24. Additionally, in vivo data suggesting the function of FBXW7 in tumor cell migration were also demonstrated. In addition, a direct link between FBXW7 and SREBP1a was established.
In conclusion, our data indicated that FBXW7 inhibits bladder cancer cells, likely through activating GSK3 phosphorylation and suppressing SREBP1a expression. Zhu et al. confirmed that p65 overexpression can inhibit the transcription of PTEN, thus stabilizing the protein expression of FBXW7, thereby increasing the ubiquitination and degradation of RhoGDIα, and ultimately promoting the migration of HUMAN BC (30). This study proved that FBXW7 inhibits the expression of SREBP1a by promoting the activation of P-GSK3β. This suggests that FBXW7 is regulated in different directions in cancer and has different effects on cancer. Therefore, this study enriched the mechanism of FBXW7’s effect on cancer, and provided new ideas for FBXW7 as a target selection.
Acknowledgments
Funding: This study was funded by grants from the Jiangxi Provincial Department of Education Project (No. GJJ180117 and GJJ150118); Science and Technology Program of Jiangxi Health and Family Planning Commission (No. 20191014, 2018B168,20175102:700890002); and Jiangxi Natural Science Foundation (No. 20202BABL206023).
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at https://tau.amegroups.com/article/view/10.21037/tau-22-146/rc
Data Sharing Statement: Available at https://tau.amegroups.com/article/view/10.21037/tau-22-146/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tau.amegroups.com/article/view/10.21037/tau-22-146/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All animal experiments were approved by the Ethics Committee of the First Affiliated Hospital of Nanchang University (approval No. 20170312), in compliance with the institutional guidelines of First Affiliated Hospital of Nanchang University for the care and use of animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Lenis AT, Lec PM, Chamie K, et al. Bladder Cancer: A Review. JAMA 2020;324:1980-91. [Crossref] [PubMed]
- Liu L, Cui J, Zhao Y, et al. KDM6A-ARHGDIB axis blocks metastasis of bladder cancer by inhibiting Rac1. Mol Cancer 2021;20:77. [Crossref] [PubMed]
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019;69:7-34. [Crossref] [PubMed]
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin 2015;65:5-29. [Crossref] [PubMed]
- Kotzka J, Knebel B, Janssen OE, et al. Identification of a gene variant in the master regulator of lipid metabolism SREBP-1 in a family with a novel form of severe combined hypolipidemia. Atherosclerosis 2011;218:134-43. [Crossref] [PubMed]
- Hao M, Lin Z, Rong H, et al. Sterol regulatory element binding protein-1: Molecular cloning, tissue distribution and gene expression level in response to nutritional regulation in mud crab, Scylla paramamosain. Biochem Biophys Res Commun 2018;505:705-11. [Crossref] [PubMed]
- Bengoechea-Alonso MT, Ericsson J. A phosphorylation cascade controls the degradation of active SREBP1. J Biol Chem 2009;284:5885-95. [Crossref] [PubMed]
- Lau AW, Fukushima H, Wei W. The Fbw7 and betaTRCP E3 ubiquitin ligases and their roles in tumorigenesis. Front Biosci (Landmark Ed) 2012;17:2197-212. [Crossref] [PubMed]
- Wang Z, Liu P, Inuzuka H, et al. Roles of F-box proteins in cancer. Nat Rev Cancer 2014;14:233-47. [Crossref] [PubMed]
- Wang X, Xu W, Chen H, et al. Astragaloside IV prevents Aβ1-42 oligomers-induced memory impairment and hippocampal cell apoptosis by promoting PPARγ/BDNF signaling pathway. Brain Res 2020;1747:147041. [Crossref] [PubMed]
- Welcker M, Clurman BE. FBW7 ubiquitin ligase: a tumour suppressor at the crossroads of cell division, growth and differentiation. Nat Rev Cancer 2008;8:83-93. [Crossref] [PubMed]
- Al-Samawi AS, Aulaqi SM. Urinary bladder cancer in yemen. Oman Med J 2013;28:337-40. [Crossref] [PubMed]
- Abdollah F, Gandaglia G, Thuret R, et al. Incidence, survival and mortality rates of stage-specific bladder cancer in United States: a trend analysis. Cancer Epidemiol 2013;37:219-25. [Crossref] [PubMed]
- Jiang WG, Sanders AJ, Katoh M, et al. Tissue invasion and metastasis: Molecular, biological and clinical perspectives. Semin Cancer Biol 2015;35:S244-75. [Crossref] [PubMed]
- Krakhmal NV, Zavyalova MV, Denisov EV, et al. Cancer Invasion: Patterns and Mechanisms. Acta Naturae 2015;7:17-28. [Crossref] [PubMed]
- Clark AG, Vignjevic DM. Modes of cancer cell invasion and the role of the microenvironment. Curr Opin Cell Biol 2015;36:13-22. [Crossref] [PubMed]
- Yeh CH, Bellon M, Nicot C. FBXW7: a critical tumor suppressor of human cancers. Mol Cancer 2018;17:115. [Crossref] [PubMed]
- Cheng Y, Li G. Role of the ubiquitin ligase Fbw7 in cancer progression. Cancer Metastasis Rev 2012;31:75-87. [Crossref] [PubMed]
- Izumi N, Helker C, Ehling M, et al. Fbxw7 controls angiogenesis by regulating endothelial Notch activity. PLoS One 2012;7:e41116. [Crossref] [PubMed]
- Li M, Ouyang L, Zheng Z, et al. E3 ubiquitin ligase FBW7α inhibits cholangiocarcinoma cell proliferation by downregulating c-Myc and cyclin E. Oncol Rep 2017;37:1627-36. [Crossref] [PubMed]
- Jiang Y, Qi X, Liu X, et al. Fbxw7 haploinsufficiency loses its protection against DNA damage and accelerates MNU-induced gastric carcinogenesis. Oncotarget 2017;8:33444-56. [Crossref] [PubMed]
- Zhao J, Wang Y, Mu C, et al. MAGEA1 interacts with FBXW7 and regulates ubiquitin ligase-mediated turnover of NICD1 in breast and ovarian cancer cells. Oncogene 2017;36:5023-34. [Crossref] [PubMed]
- Tan Y, Sangfelt O, Spruck C. The Fbxw7/hCdc4 tumor suppressor in human cancer. Cancer Lett 2008;271:1-12. [Crossref] [PubMed]
- Korphaisarn K, Morris VK, Overman MJ, et al. FBXW7 missense mutation: a novel negative prognostic factor in metastatic colorectal adenocarcinoma. Oncotarget 2017;8:39268-79. [Crossref] [PubMed]
- Akhoondi S, Lindström L, Widschwendter M, et al. Inactivation of FBXW7/hCDC4-β expression by promoter hypermethylation is associated with favorable prognosis in primary breast cancer. Breast Cancer Res 2010;12:R105. [Crossref] [PubMed]
- Sato M, Rodriguez-Barrueco R, Yu J, et al. MYC is a critical target of FBXW7. Oncotarget 2015;6:3292-305. [Crossref] [PubMed]
- Kitade S, Onoyama I, Kobayashi H, et al. FBXW7 is involved in the acquisition of the malignant phenotype in epithelial ovarian tumors. Cancer Sci 2016;107:1399-405. [Crossref] [PubMed]
- Liu F, Zou Y, Wang F, et al. FBXW7 Mutations Promote Cell Proliferation, Migration, and Invasion in Cervical Cancer. Genet Test Mol Biomarkers 2019;23:409-17. [Crossref] [PubMed]
- Liu L, Zhao X, Zhao L, et al. Arginine Methylation of SREBP1a via PRMT5 Promotes De Novo Lipogenesis and Tumor Growth. Cancer Res 2016;76:1260-72. [Crossref] [PubMed]
- Zhu J, Li Y, Chen C, et al. NF-κB p65 Overexpression Promotes Bladder Cancer Cell Migration via FBW7-Mediated Degradation of RhoGDIα Protein. Neoplasia 2017;19:672-83. Erratum in: Neoplasia 2021;23:1179-82. [Crossref] [PubMed]
(English Language Editor: J. Jones)


