
Renal cancer stem cell-derived sEVs impair renal function by inducing renal cell ERS and apoptosis in mice
Introduction
Cancer stem cells (CSCs) represent a population of stem-like cells within a tumor that are particularly endowed with the capacity of self-renewal, proliferation, and multidirectional differentiation. CSCs are considered initiating cells of tumorigenesis, tumor development, and metastasis (1). In 2005, Florek et al. discovered for the first time RCC stem cells from renal cell carcinoma cells by a cytosphere (sp) formation experiment, which confirmed the existence of renal CSCs (2). The existing studies on CSCs have been mainly focused on the occurrence and development of the tumor, but the impact of CSCs on the local environment has not been sufficiently investigated (3). Renal function impairment is the most severe effect on the local environment in kidney tumor patients, because the kidneys function as a blood filter.
Among the manifestations of renal cell carcinoma (RCC), renal function impairment has been of great concern to clinicians since it not only the reflection of tumor development and organism function (4,5), but also determines life quality and survival outcomes of patients (6-8). Renal function impairment could occur even at the early stage of RCC. However, RCC lacks specific symptoms and has a highly variable presentation, which hinders the identification of early-stage renal function impairment (9). The mechanism of RCC-induced renal function impairment is rather complicated because tumor damage could be not only physical but also biochemical.
Small extracellular vesicles (sEVs) are lipid bilayer vesicles secreted by cells. Carrying and conserving a variety of proteins, nucleic acids and cytokines, sEVs transfer biological signals between cells and exert unique regulation roles in a spectrum of physiological and pathological processes (10-13). The contents of sEVs are determined by the tissue or cells of origin, which reflects different physiological and pathological conditions (14). The RNA content of tumor-derived small extracellular vesicles (TsEVs) is similar to that of its original cells or tissues (15-17). Recent studies have shown that TsEVs exert negative influence on normal organs or tissues (18), leading to tumor progression, inflammation, and a series of other pathological changes (19-22). These findings indicated that TsEVs probably play a role of invisible tumor. It is noteworthy that sEVs released by CSCs induced angiogenesis and the premetastatic niche (3). However, whether sEVs derived from renal CSCs (RCSCs) affect renal function and its mechanism is still unknown. Considering their unique characteristics of targeting and homing (23-25), we speculated that sEVs secreted by RCSCs would specifically return to renal cells and induce a cascade of pathological changes.
In this study, we first established that RCSC-sEVs caused renal cell apoptosis and loss of renal function, which was confirmed by the changes in the 24-hour urinary protein and serum creatinine, as well as by the pathological examination of the renal tissue. Our in vitro experiments confirmed that renal RCSC-sEVs induced endoplasmic reticulum stress (ERS) in normal renal cells. Further exploration of the underlying mechanism revealed that miR-142-3p carried by RCSC-sEVs affected the expression of ERp44 and finally caused apoptosis and renal function impairment. We present the following article in accordance with the ARRIVE reporting checklist (available at https://tau.amegroups.com/article/view/10.21037/tau-21-1007/rc).
Methods
Acquisition and analysis of miRNA expression
The miRNA expression was retried from TCGA. Data analysis and prediction were conducted using Sangerbox, Oncomir (26), Diana TarBase (27), Encyclopedia of RNA Interactome (ENCORI) (28), and GEPIA (29). Details are available in the Appendix 1. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Cell culture, sEVs isolation, and identification
Human RCC cell lines A498, 786-O, OS-RC-2, ACHN, and SW839, and the human renal tubular epithelial cell line HK2 were obtained and cultured according to the instructions and previous research (30). RCSCs were isolated as described earlier (31), and their morphology was observed. A single sp was transplanted to a new T25 flask and proliferated stably. Then, RCSCs were identified by Western blot (WB) and real-time quantitative polymerase chain reaction (RT-qPCR). sEVs were isolated as previously reported, including in our earlier study (32-34). Details of the operations are described in the Appendix 1. The obtained sEVs were placed under a transmission electron microscope (TEM, Hitachi H-7650) to morphological observation and evaluation. A Nanoparticle Tracking Analyzer (Particle Metrix, Germany) was used to determine the size and distribution of the sEVs. WB was applied to detect the sEVs marker proteins.
miRNA screening
An exoRNeasy Serum/Plasma Starter Kit (QIAGEN, Germany) was used to extract sEVs total RNAs after isolation. Cell total RNAs were extracted using an RNA-Quick Purification Kit (ES science, China). A microRNA Reverse Transcription Kit (EZB-miRT2, HiFunBio, Shanghai, China) was utilized for RNA reverse transcription, and the 2 × SYBR Green qPCR Master Mix (ROX2 plus, HiFunBio, Shanghai, China) kit was used for qPCR with an ABI 7500 fast real-time fluorescent quantitative PCR instrument. The primers were designed and synthesized by RiboBio (RiboBio Biotechnology Ltd., Guangzhou, China).
Local injection of RCSC-sEVs in mice
To investigate the effect of RCSC-sEVs on renal function, we injected RCSC-sEVs into mouse kidneys. The methods are briefly introduced below.
Based on the protocol of previous studies on the effect of human EVs on kidney damage, healthy male C57 BL/6 mice were used for their human-like whole genome and physiological structure. Eight-week-old C57 mice were obtained from Shanghai JIHUI Laboratory Animal Breeding Co., Ltd. (SCXK [Shanghai, China] 2017-0012). Experiments were performed under a project license (No. SHRM-IACUC-042) granted by Institutional Animal Care and Use Committee Board of SHRM (SYXK [Shanghai, China] 2021-0007), in compliance with China national guidelines for the care and use of animals. Forty-eight male mice were randomly divided into group A and group B. Each group was randomly divided into four sub-groups, labeled as A(1)–A(4) and B(1)–B(4), respectively, with six mice in each of them. The mice were numbered and grouped by a computer. Then, they were anesthetized using 1% sodium pentobarbital (0.08 mg/g) and subcutaneously injected with butorphanol (0.001 mg/g). Next, 150 µL of liquid was injected into kidney tissue around the right renal artery in group A, whereas in group B, 150 µL of liquid was injected into kidney tissue around both renal arteries. The injection liquid had the following content: A(1), B(1): normal saline; A(2), B(2): sEVs isolated from ACHNsp (ACHNsp-sEVs), 1×1011 particles/ mL; A(3),B(3): sEVs isolated from 786Osp (786Osp-sEVs), 1×1011 particles/mL; A(4),B(4): sEVs isolated from HK2 cells (HK2-sEVs), 1×1011 particles/mL. Each mouse was injected once a week (every Monday). Every Sunday, three mice were transferred to the metabolic cage to collect 24-hour urine samples. Blood samples (100 µL) were collected from the tail vein. To observe the pathological changes, at the end of weeks 3 and 6, three mice were sacrificed by CO2, and bilateral kidneys were collected for further examination. 24-hour urinary protein was detected every week using a mouse urinary protein ELISA kit (Shanghai Jianglai Industrial Limited By Share Ltd., China). Serum creatinine (S-Cr) was measured using a mouse serum creatinine (S-Cr) Elisa kit (GTX, USA). The order of measurements and cage location were randomly decided to minimize potential confounders.
Histological and immunohistochemistry analysis
The kidneys were collected and fixed in 10% formalin for Periodic Acid-Schiff (PAS) and immunohistochemistry (IHC) staining. Tissue specimens were embedded in paraffin and sliced into 4-µm-thick sections. PAS staining was used for kidney histological observation and evaluation. IHC was employed for determining the expression of apoptosis protein. The observation was conducted using a light microscope (DM 4000B, Leica Micro systems, Buffalo Grove, IL, USA). Image-Pro Plus 6.0 software was utilized for the evaluation of protein expression.
Apoptosis examination (Western Blot, TUNEL, and Flow cytometry)
Cell slides were attached to the bottom of a 6-well plate and HK2 cells were seeded at the density of 25% and divided into three groups. (I) Control group: normally cultured; (II) ACHNsp-sEVs group: 1×1010 particles/mL ACHNsp-sEVs; (III) 786Osp-sEVs group: 1×1010 particles/mL 786Osp-sEVs. The slides were collected after 24 and 48 hours. One-step TUNEL apoptosis detection kit (Beyotime, China) was used for apoptosis detection. Image-Pro Plus 6.0 was employed to calculate the apoptosis ratio. HK2 cells were cultured and intervened under the same conditions for 24 and 48 hours. Following the use of the Annexin V-FITC/PI Cell Apoptosis Detection Kit (Servicebio®, China), cell apoptosis was examined by flow cytometry.
Application of antagomir-142-3p and miR142-3p inhibitor
In vivo, 30 6–8-week-old male C57 mice were divided into five groups. The following grouping and treatment were applied. (I) Control group: normal saline, 150 µL, local injection around renal artery, once a week; (II) ACHNsp-sEVs group: ACHNsp-sEVs, 1×1011 particles/mL, 150 µL, local injection around renal artery, once a week; (III) ACHNsp-sEVs + antagomir group: antagomir-142-3p (RIBOBIO biotechnology, Guangzhou, China), 100 nmol/kg for three days, caudal vein injection one week in advance. ACHNsp-sEVs, 1×1011 particles/mL, 150 µL, local injection around renal artery, once a week; (IV) 786Osp-sEVs group: 786Osp-sEVs, 1×1011 particles/mL, 150 µL local injection around renal artery, once a week; (V) 786Osp-sEVs + antagomir group: antagomir-142-3p, 100 nmol/kg for 3 days, caudal vein injection one week in advance. 786Osp-sEVs, 1×1011 particles/mL, 150 µL, local injection around renal artery were applied once a week. The aforementioned examinations were performed (24-hour urinary protein, serum creatinine, PAS, and IHC).
In vitro, HK2 cells were seeded in 6-well plates and divided into three groups as follows: (I) control group, (II) ACHNsp-sEVs group, and (III) 786Osp-sEVs group. Samples of each group were seeded into two plates and cultured with medium/sEVs or medium/sEVs + miR-142-3p inhibitor separately. After 24 hours of intervention, cell apoptosis and ERS were observed by WB, TUNEL staining, and FCM. The methods described above were used.
miR142-3p regulates ERp44 expression
The possible target gene of miR-142-3p was predicted by TarBase (19) and verified by dual-luciferase activity assay. Constructed pmirglo-mir-142-3p-erp44-wt plasmid and miRNA mimics were co-transfected into HEK293T cells by liposomal transcription reaction (Hieff TransTM, China). The plasmid was constructed by Shanghai Integrated Biotech Solutions Co., Ltd (IBSBIO, China). Forty-eight hours later, the dual-luciferase activities (firefly luciferase and sea kidney luciferase) were detected by a multi-mode microplate reader (SpectraMax® ID5, Molecular Devices, USA), and the cell luciferase activities were calculated.
Effect of miR-142-3p/ERp44 regulation on apoptosis/ERS
The overexpression plasmid of ERp44 was constructed by Shanghai Integrated Biotech Solutions Co., Ltd (IBSBIO, China). The following grouping and examination were conducted: (I) ERp44 nc; (II) ERp44 nc + miR-142-3p mimic; (III) ERp44 oe + miR-142-3p mimic; and (IV) ERp44 oe. Forty-eight hours after the transfection, the cells were seeded into six-well plates at a density of 30% and ACHNsp-sEVs were added into the medium (1×1010 particles/mL). Twenty-four hours later, the protein was collected for Western blot. The aforementioned interventions were repeated with cell-climbing slides for TUNEL staining.
Effect of ERp44 expression regulation and the activity of PERK in HK2
HK2 cells were seeded in six-well plates. Then, ERp44 interference plasmid (IBSBIO, China) and PERK inhibitor (GSK2606414, Sigma-Aldrich, Germany) were used. After 48 h, the cells were seeded into six-wells plates at a density of 30% and ACHNsp-sEVs were added into the medium (1×1010 particles/mL). The following groups were set: (I) ERp44 nc; (II) ERp44knock down (kd); (III) PERK inhibitor; and (IV) PERK inhibitor + ERp44 kd. The protein was collected 24 hours later for Western blot analysis. The interventions described above were repeated with cell-climbing slides for TUNEL staining.
Statistical analysis
All the data are presented as mean ± standard deviation (SD). The inter-group differences were examined using Student’s t-test. The statistical significance of the differences between groups was assessed by one-way analysis of variance (ANOVA) and Dunnett’s multiple comparison tests. Statistical differences were considered significant at P<0.05. SPSS 22.0 (SPSS, Inc., Chicago, IL, USA) was employed for data analysis.
Results
miR-142-3p is overexpressed in RCSC-sEVs
The differences in the microRNAs expression between human RCC and normal tissues are presented in Figure 1A and Table S1 Based on the survival outcomes (Figure 1B and Figure S1), miR-885-5p, miR-891a-5p, miR-155-5p, miR-875-5p, miR-1293-5p, miR-21-5p, miR-584-5p, miR-142-3p, miR-3613, miR-210-3p, miR-33b-5p, and miR-1228-5p were selected for RT-qPCR analysis. By comparing human RCC cell lines A498, 786-O, OS-RC2, ACHN, SW839, and the human renal tubular epithelial cell line HK2, we verified the expression differences (Figure 1B). The expression of miR-142-3p was higher in the sEVs derived from ACHNsp and 786Osp cells than in the adherent cells (Figure 1B).
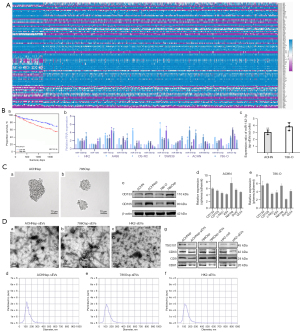
RCSCs and sEVs morphology, protein markers, and particle size
RCSC sp (ACHNsp and 786Osp) was observed after 10 days (Figure 1C). The identification of RCSCs is presented in Figure 1C, which was consistent with previous research findings (35,36). sEVs derived from ACHNsp, 786Osp and HK2 obtained by ultracentrifugation were observed under transmission electron microscope with a magnification of ×42,000. Microscopically, the typical lipid double membrane of sEVs was observed, which was a ring-like structure (Figure 1D). NTA analysis showed that the diameter of sEVs was 50–200 nm (Figure 1D). Positive Western blot results were obtained for the EVs marker proteins CD9, CD63, and CD81, and TSG101 (Figure 1D).
Bilateral kidney injection of RCSC-sEVs causes renal function impairment in mice
The RCSC-sEVs injection is depicted in Figure 2A. The sEVs are displayed in Figure 2B. The 24-hour urinary protein (Figure 2C) and serum creatinine (Figure 2C) in the mice injected with RCSC-sEVs were higher than those in the mice injected with normal saline. Notably, the renal function of the mice treated with bilateral RCSC-sEVs injection was lower than that of the ones treated with unilateral injection. We speculate that this result was obtained because a few of the sEVs reached the contralateral kidney, and the loss of unilateral renal function was compensated by the healthy kidney. When both kidneys were injected with RCSC-sEVs, the loss of renal function could not be compensated, and the 24-hour urinary protein and serum creatinine were increased. The post-experience power calculation shows that the existing inspection power was sufficient.
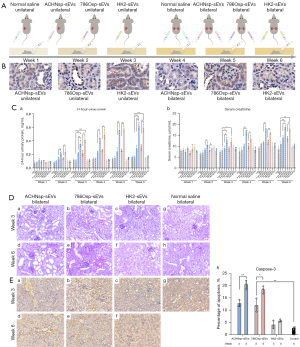
PAS staining found pathological changes in the mice injected with RCSC-sEVs, such as tubulointerstitial fibrosis, glomerular atrophy, disappearance of the brush edge of the renal tubules, bare basement membrane, atrophy, or expansion of the renal tubules (Figure 2D). The expression of caspase-3 was also higher in the mice with RCSC-sEVs injection, which indicated more apoptosis (Figure 2E).
FCM and TUNEL showed that the uptake of RCSC-sEVs induced HK2 cell apoptosis
After culturing with RCSC-sEVs (Figure 3A), apoptosis of HK2 cells was detected by FCM (Figure 3B) and TUNEL staining (Figure 3C). The results showed that the percentage of apoptotic cells significantly increased after 24 and 48 hours. This result, due to the continual RCSC-sEVs existence ensured by timely changing the medium, was confirmed by TUNEL staining.
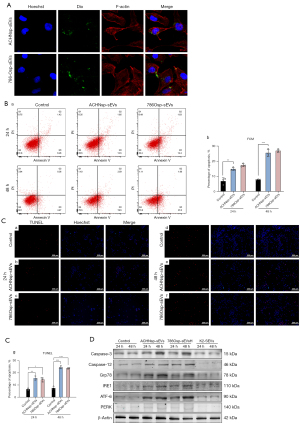
WB showed that ERS occurred in HK2 after the uptake of RCSC-sEVs
To explore the mechanisms behind cell apoptosis, related proteins were detected by WB (Figure 3D). We found that the expression of caspase-3 in HK2 cells was higher after culturing with RCSC-sEVs. Furthermore, we established that the expression of Caspase-12, a specific maker protein of apoptosis caused by ERS, increased too. We also exanimated the ERS markers ATF6, GRP78, and IRE1. The results showed that the expression levels of the three proteins were increased in different degrees, suggesting that a complicated mechanism was involved in the ERS caused by RCSC-sEVs.
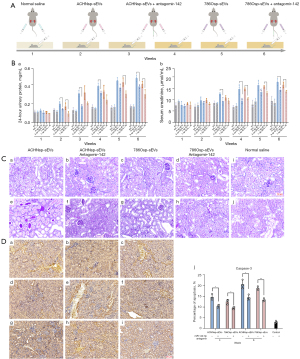
RCSC-sEVs mir-142-3p caused renal function impairment
Considering the pro-apoptotic effect of RCSC-sEVs on normal renal cells in vitro and in vivo and the high expression of miR-142-3p in RCSC-sEVs, we speculated that miR-142-3p may be involved in apoptosis. To suppress the potential effect of miR-142-3p, we first injected antagomir-142-3p into the mice (Figure 4A) and then injected RCSC-sEVs. Our results showed that the renal function impairment was attenuated using both RCSC-sEVs and antagomir-142-3p (Figure 4B). PAS staining results (Figure 4C) showed that less pathological changes occurred in the groups treated with antagomir-142-3p than in the RCSC-sEVs groups. The expression of caspase-3 in the mouse kidneys was also decreased (Figure 4D).
miR-142-3p induced ERS and apoptosis in HK2 cells
In vitro, FCM results showed that cell apoptosis was decreased after the miR-142-3p inhibitor treatment (Figure 5A). The expression of Caspase-12, GRP78, PERK, and ATF6 was also changed by the use of miR-142-3p as a mimic or inhibitor (Figure 5B). TUNEL results also showed lower percentages of apoptotic cells in both the AHCNsp-sEVs and 786Osp-sEVs groups after the treatment with the miR-142-3p inhibitor (Figure 5C).
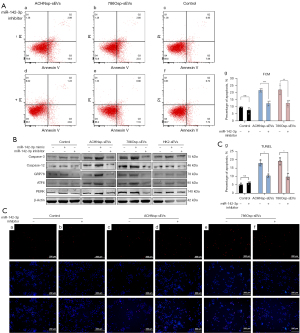
miR-142-3p affected ERp44 expression
According to data available on DIANA-TarBase v8 (27) and existing research (37), ERp44 is a possible target gene of miR-142-3p (Figure 6A) that affected the survival outcome of the patients (Figure 6A). The dual-luciferase reporter gene detection showed significantly decreased fluorescence intensity of the miR-142-3p mimic and ERp44-wt co-transfection group, whereas no significant changes in the fluorescence intensity of miR-142-3p mimic and ERp44-mut were found in the co-transfection group (Figure 6A).
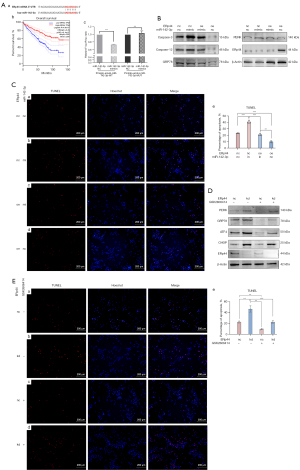
ERp44 overexpression decreased the expression of ERS proteins and cell apoptosis
The expression levels of caspase-3 and caspase-12 in HK2 cultured with RCSC-sEVs were decreased by the overexpression of ERp44. The expression of the ERS proteins GRP78 and PERK was also decreased. However, this decrease was alleviated by the application of miR-142-3p mimic (Figure 6B). TUNEL staining of the same treatment also showed that the overexpression of ERp44 reduced the apoptosis in HK2 cells cultured with RCSC-sEVs (Figure 6C).
ERp44 knockdown increased the expression of PERK in HK2 cells
ERp44 knockdown improved the expression of PERK and GRP78 in the PERK-CHOP pathway. WB results showed that ERp44 knockdown in HK2 cells cultured with RCSC-sEVs caused higher level of PERK, GRP78, ATF4, and CHOP expression than those of the control group (nc), suggesting that the PERK-CHOP pathway of ERS was activated. Meanwhile, the application of PERK inhibitors effectively reduced the expression of the aforementioned proteins (Figure 6D). TUNEL staining of the same treatment also revealed that the percentage of apoptotic cells was higher in ERp44-kd cells, but decreased when using the PERK inhibitor (Figure 6E).
Discussion
Renal function is closely related to making a clinical decision for RCC treatment and to patients’ postoperative survival outcomes. Systematic evaluation of the renal function of RCC patients is an important and interdisciplinary work involving knowledge of nephrology, urology, and oncology, which demanding and challenging to urologists. In the early stages of RCC, the impairments of renal function are usually not obvious because of the occult symptoms and the compensatory effect of the contralateral kidney. It is noteworthy that obvious pathological changes in most patients are usually observed in the normal renal tissue adjacent to the RCC tissue. However, few studies have investigated the relationship between such changes and the occurrence of renal function impairment. Tumorigenesis, tumor development, and metastasis are considered to be due to the development and spread of CSCs. Hence, we speculate that RCSCs induce the pathological changes in the adjacent renal tissue. Considering the characteristics of homing, RCSC-sEVs may return to the normal renal cells through the paracrine pathway, transmit specific biological signals, and affect normal renal cells.
In this study, renal function impairment and renal cell apoptosis were observed after the administration of a local injection of RCSC-sEVs. According to the existing theory, three pathways are involved in mammalian cell apoptosis: extrinsic pathway, mitochondrial pathway, and the ERS pathway which is typically marked by increased caspase-12 expression. In this study, we found increased caspase-12 was in the apoptotic cells, and thus it was possible that ERS was caused by RCSC-sEVs, which induced renal cell apoptosis. Accordingly, three pathways were involved in ERS apoptosis: ATF6-CHOP, IRE1-CHOP, and PERK-CHOP (38). Although all these three pathways could induce ERS and apoptosis, the activation of the PERK-eIF2α-ATF4 pathway is a prerequisite for CHOP expression (39), which was also found to be upregulated in this study. The C/EBP homologous protein (CHOP), also known as GADD153, is a transcription factor activated at multiple levels during ERS and a critical cause of cell apoptosis (40). When unfolded protein reaction (UPR) occurs, PERK activates eIF2α, leading to temporary translational repression, whereas CHOP could activate growth arrest and DNA damage inducible gene 34 (GADD34) and aggravate UPR. Meanwhile, CHOP could activate ERO1α and lead to accumulation of reactive oxygen in cells and aggravation of protein misfolding of (41), accelerating cell death.
In the present study, we found that the PERK-CHOP pathway was activated by ERp44 knockdown. ERp44 encodes a member of the protein disulfide isomerase (PDI) family of endoplasmic reticulum proteins and is involved in early protein synthesis in the endoplasmic reticulum (42). Existing evidence has shown that ERp44 downregulation reduces the release of internal Ca2+ and disrupts its homeostasis in the endoplasmic reticulum lumen, subsequently causing endoplasmic reticulum dysfunction and ERS (43-45).
The regulation of ERp44 is multifaceted. In this study, we established that miR-142-3p, carried by RCSC-sEVs, decreased ERp44 expression. As carriers of biological signals, sEVs have a double-layer membrane structure which protects RNAs from degradation by RNase and transports them to recipient cells (46), thereby exerting an important regulatory role in tumor occurrence and development (47). Earlier studies showed that the expression of miR-142-3p was upregulated in renal cell carcinoma and inhibited the proliferation and migration of normal cells (48,49). Additionally, RCC patients with higher expression of miR-142-3p usually have bleak prognosis (50). Furthermore, the expression of miR-142-3p in urine and peripheral blood samples of patients who received kidney transplantation was upregulated, which usually indicates the presence of acute kidney injury, interstitial fibrosis, and renal tubular atrophy (51,52). The results of this study are indirectly consistent with those of the aforementioned research, meaning miR-142-3p could act as an external biological signal to induce stress in normal renal cells, resulting in a series of pathological changes.
In conclusion, RCSC-sEVs inhibit the expression of ERp44 by delivering miR-142-3p, thus inducing continuous ERS and accelerating apoptosis through the PERK-CHOP pathway. The ultimate effect these processes is renal function impairment.
However, the integral mechanism of damage exerted by RCSC-sEVs on renal function is rather complicated, which will be explored further in our future research.
Conclusions
The present study evidences that the loss of renal function in RCC patients might be induced not only by tumor development, but also by renal cell apoptosis caused by RCSC-sEVs. As a natural vector of miR-142-3p, RCSC-sEVs inhibit the expression of ERp44 and induce renal cell apoptosis caused by ERS, thereby leading to renal function impairment.
Acknowledgments
The authors express their gratitude to the Central Laboratory of Shanghai, 10th Peoples’ Hospital for the service and assistance they provided during the study.
Funding: This work was financially supported by the National Nature Science Foundation of China (Project Nos. 81972393, 81772705 and 31570775).
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at https://tau.amegroups.com/article/view/10.21037/tau-21-1007/rc
Data Sharing Statement: Available at https://tau.amegroups.com/article/view/10.21037/tau-21-1007/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tau.amegroups.com/article/view/10.21037/tau-21-1007/coif). JZ reports that this work was supported by National Nature Science Foundation of China (Project Nos. 81972393, 81772705 and 31570775). The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Eight-week-old C57 mice were obtained from Shanghai JIHUI Laboratory Animal Breeding Co., Ltd. (SCXK [Shanghai, China] 2017-0012). Experiments were performed under a project license (No. SHRM-IACUC-042) granted by Institutional Animal Care and Use Committee Board of SHRM (SYXK [Shanghai, China] 2021-0007), in compliance with China national guidelines for the care and use of animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Clarke MF, Dick JE, Dirks PB, et al. Cancer stem cells--perspectives on current status and future directions: AACR Workshop on cancer stem cells. Cancer Res 2006;66:9339-44. [Crossref] [PubMed]
- Florek M, Haase M, Marzesco AM, et al. Prominin-1/CD133, a neural and hematopoietic stem cell marker, is expressed in adult human differentiated cells and certain types of kidney cancer. Cell Tissue Res 2005;319:15-26. [Crossref] [PubMed]
- Grange C, Tapparo M, Collino F, et al. Microvesicles released from human renal cancer stem cells stimulate angiogenesis and formation of lung premetastatic niche. Cancer Res 2011;71:5346-56. [Crossref] [PubMed]
- Betjes MG, Litjens NH. Chronic kidney disease and premature ageing of the adaptive immune response. Curr Urol Rep 2015;16:471. [Crossref] [PubMed]
- Kato S, Chmielewski M, Honda H, et al. Aspects of immune dysfunction in end-stage renal disease. Clin J Am Soc Nephrol 2008;3:1526-33. [Crossref] [PubMed]
- Jeon HG, Gong IH, Hwang JH, et al. Prognostic significance of preoperative kidney volume for predicting renal function in renal cell carcinoma patients receiving a radical or partial nephrectomy. BJU Int 2012;109:1468-73. [Crossref] [PubMed]
- Antonelli A, Minervini A, Sandri M, et al. Below Safety Limits, Every Unit of Glomerular Filtration Rate Counts: Assessing the Relationship Between Renal Function and Cancer-specific Mortality in Renal Cell Carcinoma. Eur Urol 2018;74:661-7. [Crossref] [PubMed]
- Shingarev R, Jaimes EA. Renal cell carcinoma: new insights and challenges for a clinician scientist. Am J Physiol Renal Physiol 2017;313:F145-54. [Crossref] [PubMed]
- Gudbjartsson T, Thoroddsen A, Petursdottir V, et al. Effect of incidental detection for survival of patients with renal cell carcinoma: results of population-based study of 701 patients. Urology 2005;66:1186-91. [Crossref] [PubMed]
- Raposo G, Nijman HW, Stoorvogel W, et al. B lymphocytes secrete antigen-presenting vesicles. J Exp Med 1996;183:1161-72. [Crossref] [PubMed]
- Lai RC, Chen TS, Lim SK. Mesenchymal stem cell exosome: a novel stem cell-based therapy for cardiovascular disease. Regen Med 2011;6:481-92. [Crossref] [PubMed]
- Peters PJ, Geuze HJ, Van der Donk HA, et al. Molecules relevant for T cell-target cell interaction are present in cytolytic granules of human T lymphocytes. Eur J Immunol 1989;19:1469-75. [Crossref] [PubMed]
- Wolfers J, Lozier A, Raposo G, et al. Tumor-derived exosomes are a source of shared tumor rejection antigens for CTL cross-priming. Nat Med 2001;7:297-303. [Crossref] [PubMed]
- Clayton A, Mason MD. Exosomes in tumour immunity. Curr Oncol 2009;16:46-9. [Crossref] [PubMed]
- H Rashed M. Exosomes: From Garbage Bins to Promising Therapeutic Targets. Int J Mol Sci 2017;18:538. [Crossref] [PubMed]
- Soung YH, Ford S, Zhang V, et al. Exosomes in Cancer Diagnostics. Cancers (Basel) 2017;9:8. [Crossref] [PubMed]
- Rabinowits G, Gerçel-Taylor C, Day JM, et al. Exosomal microRNA: a diagnostic marker for lung cancer. Clin Lung Cancer 2009;10:42-6. [Crossref] [PubMed]
- Keklikoglou I, Cianciaruso C, Güç E, et al. Chemotherapy elicits pro-metastatic extracellular vesicles in breast cancer models. Nat Cell Biol 2019;21:190-202. [Crossref] [PubMed]
- Shang A, Gu C, Wang W, et al. Exosomal circPACRGL promotes progression of colorectal cancer via the miR-142-3p/miR-506-3p- TGF-β1 axis. Mol Cancer 2020;19:117. [Crossref] [PubMed]
- Xie M, Yu T, Jing X, et al. Exosomal circSHKBP1 promotes gastric cancer progression via regulating the miR-582-3p/HUR/VEGF axis and suppressing HSP90 degradation. Mol Cancer 2020;19:112. [Crossref] [PubMed]
- Hou PP, Luo LJ, Chen HZ, et al. Ectosomal PKM2 Promotes HCC by Inducing Macrophage Differentiation and Remodeling the Tumor Microenvironment. Mol Cell 2020;78:1192-1206.e10. [Crossref] [PubMed]
- Jiang F, Chen Q, Wang W, et al. Hepatocyte-derived extracellular vesicles promote endothelial inflammation and atherogenesis via microRNA-1. J Hepatol 2020;72:156-66. [Crossref] [PubMed]
- Perets N, Betzer O, Shapira R, et al. Golden Exosomes Selectively Target Brain Pathologies in Neurodegenerative and Neurodevelopmental Disorders. Nano Lett 2019;19:3422-31. [Crossref] [PubMed]
- Batrakova EV, Kim MS. Using exosomes, naturally-equipped nanocarriers, for drug delivery. J Control Release 2015;219:396-405. [Crossref] [PubMed]
- van den Boorn JG, Dassler J, Coch C, et al. Exosomes as nucleic acid nanocarriers. Adv Drug Deliv Rev 2013;65:331-5. [Crossref] [PubMed]
- Wong NW, Chen Y, Chen S, et al. OncomiR: an online resource for exploring pan-cancer microRNA dysregulation. Bioinformatics 2018;34:713-5. [Crossref] [PubMed]
- Karagkouni D, Paraskevopoulou MD, Chatzopoulos S, et al. DIANA-TarBase v8: a decade-long collection of experimentally supported miRNA-gene interactions. Nucleic Acids Res 2018;46:D239-45. [Crossref] [PubMed]
- Navarro-Yepes J, Burns M, Anandhan A, et al. Oxidative stress, redox signaling, and autophagy: cell death versus survival. Antioxid Redox Signal 2014;21:66-85. [Crossref] [PubMed]
- Tang Z, Li C, Kang B, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 2017;45:W98-W102. [Crossref] [PubMed]
- Kornilov R, Puhka M, Mannerström B, et al. Efficient ultrafiltration-based protocol to deplete extracellular vesicles from fetal bovine serum. J Extracell Vesicles 2018;7:1422674. [Crossref] [PubMed]
- Khan MI, Czarnecka AM, Helbrecht I, et al. Current approaches in identification and isolation of human renal cell carcinoma cancer stem cells. Stem Cell Res Ther 2015;6:178. [Crossref] [PubMed]
- Kosaka N, Yoshioka Y, Hagiwara K, et al. Functional analysis of exosomal microRNA in cell-cell communication research. Methods Mol Biol 2013;1024:1-10. [Crossref] [PubMed]
- Montecalvo A, Larregina AT, Morelli AE. Methods of analysis of dendritic cell-derived exosome-shuttle microRNA and its horizontal propagation between dendritic cells. Methods Mol Biol 2013;1024:19-40. [Crossref] [PubMed]
- Wu R, Huang C, Wu Q, et al. Exosomes secreted by urine-derived stem cells improve stress urinary incontinence by promoting repair of pubococcygeus muscle injury in rats. Stem Cell Res Ther 2019;10:80. [Crossref] [PubMed]
- Bussolati B, Dekel B, Azzarone B, et al. Human renal cancer stem cells. Cancer Lett 2013;338:141-6. [Crossref] [PubMed]
- Qu L, Wu Z, Li Y, et al. A feed-forward loop between lncARSR and YAP activity promotes expansion of renal tumour-initiating cells. Nat Commun 2016;7:12692. [Crossref] [PubMed]
- Gottwein E, Corcoran DL, Mukherjee N, et al. Viral microRNA targetome of KSHV-infected primary effusion lymphoma cell lines. Cell Host Microbe 2011;10:515-26. [Crossref] [PubMed]
- Fung TS, Liu DX. Coronavirus infection, ER stress, apoptosis and innate immunity. Front Microbiol 2014;5:296. [Crossref] [PubMed]
- Fels DR, Koumenis C. The PERK/eIF2alpha/ATF4 module of the UPR in hypoxia resistance and tumor growth. Cancer Biol Ther 2006;5:723-8. [Crossref] [PubMed]
- Li Y, Guo Y, Tang J, et al. New insights into the roles of CHOP-induced apoptosis in ER stress. Acta Biochim Biophys Sin (Shanghai) 2015;47:146-7. [Crossref] [PubMed]
- Marciniak SJ, Yun CY, Oyadomari S, et al. CHOP induces death by promoting protein synthesis and oxidation in the stressed endoplasmic reticulum. Genes Dev 2004;18:3066-77. [Crossref] [PubMed]
- Watanabe S, Harayama M, Kanemura S, et al. Structural basis of pH-dependent client binding by ERp44, a key regulator of protein secretion at the ER-Golgi interface. Proc Natl Acad Sci U S A 2017;114:E3224-32. [Crossref] [PubMed]
- Chang Y, Wu Y, Liu W, et al. Knockdown of ERp44 leads to apoptosis via activation of ER stress in HeLa cells. Biochem Biophys Res Commun 2015;463:606-11. [Crossref] [PubMed]
- Ludtke SJ, Fan G, Baker ML, et al. IP3R1 - Assessing Map Interpretability at Near Atomic Resolution. Microscopy & Microanalysis 2015;21:543-4. [Crossref]
- Higo T, Hattori M, Nakamura T, et al. Subtype-specific and ER lumenal environment-dependent regulation of inositol 1,4,5-trisphosphate receptor type 1 by ERp44. Cell 2005;120:85-98. [Crossref] [PubMed]
- Xiang X, Poliakov A, Liu C, et al. Induction of myeloid-derived suppressor cells by tumor exosomes. Int J Cancer 2009;124:2621-33. [Crossref] [PubMed]
- Rak J, Guha A. Extracellular vesicles--vehicles that spread cancer genes. Bioessays 2012;34:489-97. [Crossref] [PubMed]
- Jung M, Mollenkopf HJ, Grimm C, et al. MicroRNA profiling of clear cell renal cell cancer identifies a robust signature to define renal malignancy. J Cell Mol Med 2009;13:3918-28. [Crossref] [PubMed]
- Li Y, Chen D, Jin LU, et al. Oncogenic microRNA-142-3p is associated with cellular migration, proliferation and apoptosis in renal cell carcinoma. Oncol Lett 2016;11:1235-41. [Crossref] [PubMed]
- Peng X, Pan X, Liu K, et al. miR-142-3p as a novel biomarker for predicting poor prognosis in renal cell carcinoma patients after surgery. Int J Biol Markers 2019;34:302-8. [Crossref] [PubMed]
- Domenico TD, Joelsons G, Montenegro RM, et al. Upregulation of microRNA 142-3p in the peripheral blood and urinary cells of kidney transplant recipients with post-transplant graft dysfunction. Braz J Med Biol Res 2017;50:e5533. [Crossref] [PubMed]
- Zununi Vahed S, Poursadegh Zonouzi A, Ghanbarian H, et al. Upregulated Expression of Circulating MicroRNAs in Kidney Transplant Recipients With Interstitial Fibrosis and Tubular Atrophy. Iran J Kidney Dis 2017;11:309-18. [PubMed]


