
Evaluation of NOTCH family genes’ expression and prognostic value in prostate cancer
Introduction
Prostate cancer (PCa) is one of the most prevalent tumors globally. It is predicted that there will be 268,490 newly diagnosed cases and approximately 34,500 deaths from PCa in the USA in 2022 (1). With the progress in sequencing technology, whole-genome sequencing and exome sequencing of numerous organisms have identified diverse somatic mutation patterns and pathway alterations in PCa (2-4). Recent results revealed major frequent mutations in PCa, including the androgen receptor (AR), SPOP, CHD1, and TP53, thus dividing patients with different mutations into different prognostic subtypes of PCa (5-7). Carrying different mutations may affect a patient’s sensitivity to different treatments, underlining the importance of individualized medication. A study has highlighted hormone resistance dependent or independent on the AR signaling pathway in patients with metastatic castration-resistant prostate cancer (CRPC) (8). However, in terms of clinical application, the current findings cannot meet the complex clinical requirements, and lots of drugs designed to overcome hormone resistance are now in stages of clinical trials. There is an urgent need to find the next potential biomarkers for prognosis and reaction indicators for these new therapeutic agents and targeted drugs in PCa patients.
The NOTCH family includes four distinct NOTCH receptor subtypes, namely NOTCH1, NOTCH2, NOTCH3, and NOTCH4. And for the NOTCH family genes, there are five ligands, Jagged1, Jagged2, Delta-like ligand 1 (DLL1), DLL3, and DLL4, which together make up the NOTCH signaling pathway (9). NOTCH signaling is activated by a ligand binding to receptors on adjacent cells. The binding of the receptor and ligand leads to cleavage of the NOTCH gene (10). The cleaved intracellular part translocates to the nucleus, where it cooperates with downstream target genes to influence transcription (11). The NOTCH signaling pathway plays a role in maintaining homeostasis, so its changes may cause a series of abnormal responses in the body that then lead to diseases (12).
It is worth noting that NOTCH signaling is one of the highest-ranking pathways enriched in PCa patients with a high tumor burden (13). Studies have shown that down-regulated NOTCH signaling promotes the invasion and non-anchoring growth of prostate cancer cells, and promote tumor growth and metastasis in vivo, but others elucidated that NOTCH signaling promotes tumor growth and development by triggering the AKT, FoxM1, and other targets (14-16). The exact role of NOTCH signaling in the development of PCa has not been determined because mutation-induced excessive activation of NOTCH family genes is multifaceted nature, both promoting and inhibiting cancer development. In addition, some studies have shown that the NOTCH signaling pathway can cooperate with some chemotherapy drugs (e.g., docetaxel) or anti-androgen drugs (e.g., enzalutamide) to exert synergism and an anti-tumor effect (17,18).
Although previous studies have partially determined the general expression profile and function of some NOTCH genes in PCa, there is not scientific consensus about the global landscape of NOTCH family genes in PCa and the feasibility of NOTCH family genes as potential therapeutic targets or prognostic biomarkers for PCa patients. Based on existing studies, in this study, we analyzed multiple databases and revealed that NOTCHs expressions in PCa were associated with worse disease-free survival, and there was a significant positive correlation between NOTCHs and androgen receptor. Furthermore, NOTCHs expression in PCa were associated with immune cell infiltration and NOTCHs mutation status might be a potential therapeutic target for -tinib antineoplastic drugs. We hope this could make a further explanation of NOTCH family genes’ role in PCa based on existing studies and provide a new reference for clinical prognosis and drug selection. We present the following article in accordance with the STREGA reporting checklist (available at https://tau.amegroups.com/article/view/10.21037/tau-22-281/rc).
Methods
RNA extraction and real-time quantitative reverse transcription polymerase chain reaction (qRT-PCR) assay
As for qRT-PCR, total RNA was extracted from 24 prostate cancer tissues and adjacent normal tissue using the RNeasy reagent (QIAGEN, Shanghai, China) according to the manufacturer’s instructions. The ND-2000 Nanodrop system (Thermo Scientific, USA) was used to detect the concentration and purity of RNA, and an A260/280 ratio between 1.8 and 2.0 was considered the premise of acceptable quality. Single-stranded cDNA was generated from 1 µg total RNA in a 20-µL reaction volume using oligodT primers according to the protocol supplied with the Primer Script TM RT Reagent (TaKaRa, Japan). The relative expression levels were measured by qPCR using the ABI 7900HT instrument (Applied Biosystems, USA) in a total volume of 10 µL with the SYBR green detection system (Takara, Japan) and GAPDH was used as an endogenous control. The cDNA was amplified by PCR using the following primers: NOTCH1 forward primer: 5'-TGGACCAGATTGGGGAGTTC-3', reverse primer: 5'-GCACACTCGTCTGTGTTGAC-3'; NOTCH2 forward primer: 5'-CCTTCCACTGTGAGTGTCTGA-3', reverse primer: 5'-AGGTAGCATCATTCTGGCAGG-3'; NOTCH3 forward primer: 5'-CGTGGCTTCTTTCTACTGTGC-3', reverse primer: 5'-CGTTCACCGGATTTGTGTCAC-3; NOTCH4 forward primer: 5'-TGTGAACGTGATGTCAACGAG-3', reverse primer: 5'-ACAGTCTGGGCCTATGAAACC-3'. GAPDH (internal control) forward primer: 5'-GGAGCGAGATCCCTCCAAAAT-3', reverse primer: 5'-GGCTGTTGTCATACTTCTCATGG-3'.
Immunohistochemistry (IHC)
A total of 15 PCa tissues and 6 benign prostate hyperplasia (BPH) tissues were collected from patients undergoing radical prostatectomy or transurethral prostatectomy in The Third Affiliated Hospital of Sun Yat-sen University. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study involving human tissue was approved by the Ethics Committee of The Third Affiliated Hospital of Sun Yat-sen University (No. 2019-02-153-01). All patients signed an informed consent form and agreed to the use of the surgically removed tissue. Prostate tissue samples were embedded in paraffin, which was removed with xylene, and then hydrated with ethanol. Next, 0.3% hydrogen peroxide (H2O2) and citric acid buffer (pH 6.0) were used to block endogenous peroxidase and repair antigen. Antibodies against NOTCH1 (Proteintech #20687-1-AP), NOTCH2 (Proteintech #28580-1-AP), NOTCH3 (Proteintech #55114-1-AP), and NOTCH4 (Abclonal #A8303) were incubated overnight at 4 ℃. All immunostaining scores were independently confirmed by two pathologists who were unaware of the patient's clinical information. Each pathologist took three photographs for a total of six photos were taken of each sample. Three pictures were randomly selected for final analysis.
TIMER
Tumor Immune Estimation Response (TIMER; https://cistrome.shinyapps.io/timer) is a web server for comprehensive analysis of pan-cancer gene expression patterns and tumor-infiltrating immune cells, which pre-calculates the levels of six tumor-infiltrating immune subgroups (19). In our study, the DiffExp module was used to study the differential expression between tumor and adjacent normal tissues for NOTCH family genes across all TCGA (The Cancer Genome Atlas) tumors. The gene module was used to visualize the correlation of NOTCH family genes’ expression with the level of infiltration of six types of immune cells (B cells, CD8+ T cells, CD4+ T cells, macrophages, neutrophils, and dendritic cells) by Spearman correlation in diverse cancer types.
cBioPortal
cBioPortal for Cancer Genomics (cBioPortal, http://www.cbioportal.org, version v3.2.11) is an open-access online tool integrating the raw data from large-scale genomic projects including but not limited to TCGA and ICGC (20). In this study, according to the cBioPortal online instructions (https://www.cbioportal.org/tutorials#webinar-1), the website was used for visualization and comparison of genetic alterations of NOTCH family genes in varies cancer types, including PCa. Co-occurrence and mutual exclusivity of genetic alterations between each enquired NOTCH gene were determined by log2 odds ratio, P value, and q value, and results with q value <0.05 were considered significant.
GEPIA2
Gene Expression Profiling Interacting Analysis (GEPIA2; http://gepia2.cancer-pku.cn/) is an updated version of GEPIA, which was developed by a Peking University project team and is capable of analyzing the RNA sequencing expression data of 9,736 tumors and 8,587 normal samples from TCGA and GTEx projects (21). In our study, GEPIA’s “Expression DIY” module was used to analyze the differential expression of the NOTCH family between tumor and normal tissues. We used the “PRAD” dataset, using GEPIA’s “multi-gene comparison” module, to perform a multi-gene comparative analysis of NOTCH genes. The P value cutoff value was 0.05. Student’s t-test was used to generate a P value for expression or pathological staging analysis. The “Survival Analysis” module outputs a Kaplan-Meier curve to show the relationship between NOTCH signaling and survival prognosis.
UALCAN
UALCAN (http://ualcan.path.uab.edu/) is a comprehensive, reliable, and interactive cancer omics data analysis network resource. It is built based on TCGA, MET500, and CPTAC and uses JavaScript and CSS to provide high-quality graphics (22). We used the TCGA analysis module UALCAN and selected the prostate cancer analysis module to analyze the correlation between different Gleason scores and NOTCH family genes across PCa and normal tissues, as well as in different tumor subsets based on tumor grade, sex, cancer stages and other clinicopathological characteristics.
GeneMANIA
GeneMANIA (http://www.genemania.org) is an online analytical tool that provides information on protein and genetic interactions, gene enrichment and co-expression analysis, and prediction of the function of interesting genes (23). We selected the target species as Homo sapiens on GeneMANIA’s home page, and we typed in the target gene lists that we wanted to analyze (i.e., NOTCH family genes) and the generated visual genes network was analyzed and predicted according to the physical interactions strength, co-expression relationships, prediction results, co-localization, and other indexes.
STRING
STRING (https://string-db.org/cgi) integrates public data and analyzes protein-protein interactions (PPI), including direct (physical) and indirect (functional) connections (24). We used STRING to analyze the PPI relationships between the different genes screened in the GEO database that were related to the NOTCH pathway. In brief, the identified prostate cancer-related targets were input into the STRING interaction database together with the selected Homo sapiens category for visualization of the interaction network. The confidence of the interactive network construction is 0.4–0.7, which is medium. On this basis, the K-means algorithm was used to classify the PPI networks and identify different clustering networks.
GDSC
Genomics of Drug Sensitivity in Cancer (GDSC; https://www.cancerrxgene.org/) is an online resource for therapeutic biomarker discovery in multiple cancer cells (25). In this study, we used the GDSC database to identify potential therapeutic compounds that were sensitive for the NOTCH1–2 mutation. We then constructed a volcano plot and a scatter diagram to annotate our target of interest and finally, we performed a Mann-Whitney-Wilcoxon (MWW) analysis to explore potential applications.
Statistical analysis
Gene expression data from the TCGA and GTEx databases were analyzed using Student’s t-test. The correlation analysis was evaluated using Spearman’s correlation analysis. Cox regression analyses were used to evaluate prognostic factors. All statistical examinations were performed by database-derived tools. Scatter plots and histograms were generated using GraphPad Prism 8.0. IBM SPSS 21.0 software was used for statistical analysis. The differences between groups were compared using standard Student’s t-test, paired t-test, or Mann-Whitney U test. ANOVA analysis or Kruskal-Wallis test was used to compare differences between two or more groups. P values <0.05 were statistically significant.
Results
Abnormal expression of NOTCH family genes in PCa patients
To determine the differences in the expression of NOTCH family genes in tumors and normal tissues, we used the TIMER DifExp module to analyze the mRNA levels of NOTCH1–4 in various cancer types. Consistent with a remarkable difference of NOTCH1–4 expression in breast cancer, kidney cancer, or lung cancer, significant depression of NOTCH1 and NOTCH4 was observed between PCa tissue and normal tissue (P=1.96e-9 and 2.04e-06, for each). However, NOTCH2 and NOTCH3 did not show this expression difference in PCa (P=8.20e-2 and 2.39e-1, respectively) (Figure 1A). To further verify the NOTCH family genes’ expression in PCa, we performed a screening and analysis via the TCGA-based UALCAN database and validated that NOTCH1 and NOTCH4 mRNA expression were indeed lower in PCa tissue than in normal tissue (P=1.047e-6 and 5.93e-5, respectively) (Figure 1B). IHC of the prostate specimens showed that NOTCH1, NOTCH3, and NOTCH4 levels were higher in cancer tissues than in BPH tissues, but no such phenomenon was found for NOTCH2 (Figure 1C). The following RT-qPCR of NOTCH1–4 expression was carried out. A total of 24 paired samples of PCa tissue and adjacent normal prostate tissue were collected and tested. The RT-qPCR results were essentially in accordance with the results from the online databases, NOTCH1, NOTCH3, and NOTCH4 showed a significant downtrend of transcription level in tumor tissues compared with normal tissues, but there was little difference of NOTCH2 mRNA level between tumor and normal tissues (Figure 1D). These results indicated that the suppression of the NOTCH family genes in PCa tissues may be related to tumorigenesis.
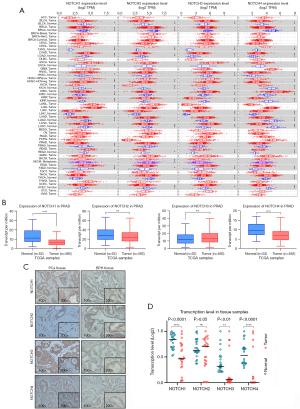
Upregulation of NOTCH family genes correlated with Gleason score and patients survival in PCa
Our analysis showed a significantly lower expression of NOTCH family genes in PCa tissue than in normal tissue, so we decided to analyze the NOTCH family genes’ transcription level alterations during PCa progression. Interestingly, the expression and clinical correlation analysis via the UALCAN database also revealed that the transcription levels of NOTCH3 and NOTCH4 were significantly enhanced with an increase of Gleason score. Compared with the patients in the Gleason score 6–8 groups, both NOTCH3 and NOTCH4 displayed the highest transcription levels in the Gleason score 9 group (Figure 2A). The positive correlation between NOTCH3–4 expression and Gleason score might indicate a promoting role of NOTCH signaling in PCa.
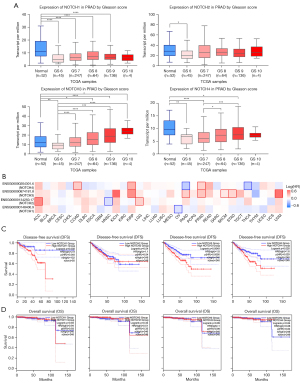
Furthermore, to evaluate the role of NOTCH family genes’ expression in the prognosis of PCa patients, we used GEPIA to assess the correlation between NOTCH1–4 expression and clinical outcomes. A survival map, which represents the survival contribution of NOTCH1–4 in multiple cancer types, showed that NOTCH3 and NOTCH4 may affect prognosis in PCa patients (Figure 2B). The disease-free survival (DFS) curve shown in Figure 2C illustrates that patients with higher transcription levels of NOTCH1, NOTCH3 and NOTCH4 had significantly poorer DFS (P=4.8e-2, 5.6e-3 and 2.9e-2, respectively). At the same time, an overall survival (OS) analysis implied that NOTCH1–4 transcription did not affect the OS of PCa patients (P=9.3e-1, 3.3e-1, 9.9e-1, and 8.4e-1, respectively, Figure 2D). In summary, NOTCH1, NOTCH3, and NOTCH4 perhaps worsen prostate cancer outlook.
Co-expression and interaction analysis of NOTCH family genes in PCa
The correlation heat map showed the internal relationships among NOTCH1–4 and the interaction between NOTCH1–4 and NOTCH-related genes (Figure 3A). As the plot shows, there was a significant moderate to high correlation among NOTCH1–4 co-expressions. The association between NOTCH1–4 and the classical NOTCH downstream genes (i.e., DLL1, DLL3, JAG1, HES1) was also verified as a generally positive correlation. Because it has been reported that there is an interaction between the NOTCH and AR signaling pathways (26,27), we screened the interaction among NOTCH1–4, AR, and AR-related genes (KLK3 and TMPRSS2). Interestingly, NOTCH1–4 and AR were observed to have a significant positive correlation, whereas AR-related KLK3 and TMPRSS2 were negatively correlated to NOTCH1–4 at the transcription level (Figure 3A). All the correlations of the genes were plotted into a scatterplot using the GEPIA database, and the significant results are presented in Figures S1,S2. We presume that it is the crosstalk between NOTCH signaling and AR that leads to the Gleason score increasing and the poor prognosis of PCa patients.
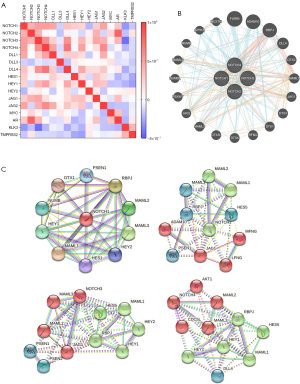
Next, to explore the potential mechanism of NOTCH family genes’ involvement in cancer, we used GeneMANIA and STRING to construct NOTCH family genes-related PPI networks. As shown in Figure 3B,3C, NOTCH1–4 had a strong physical interaction with FURIN and this correlation was further confirmed by co-expression analysis through GEPIA (Figure S3). FURIN has been reported to play a critical role in both inflammation and tumorigenesis (28,29). These results suggested that NOTCH genes might be involved in tumor promotion and inflammation, and then accelerate tumor progression through AR signaling and FURIN.
Effect of NOTCH family genes’ mutations on tumor-related biological pathways in PCa
We conducted a comprehensive analysis to uncover the molecular characteristics and genetic alteration of NOTCH family genes using the online dataset cBioportal. The results showed that NOTCH1, NOTCH2, NOTCH3, and NOTCH4 had 7%, 8%, 5%, and 5% mutational frequency in the queried PCa samples, respectively. (Figure 4A). Among these mutations, Shallow Deletion and Amplification accounted for the highest proportion (Figure S4). Mutations are always accompanied by alteration of some other genes. The NOTCH-mutation-related differentially expressed genes (DEGs) in PCa were collected and the top 10 most significant DEGs of NOTCH1–4 are shown in Figure 4B. At the same time, the common genes among NOTCH1–4 mutation-related DEGs were identified (Figure 4C, Table 1). Armed with this data, GO enrichment analysis and the network of enriched terms were performed to explore the pathway(s) affected by NOTCH1–4 mutations. As shown in Figure 4D,4E and Table 2, the functions of NOTCH signaling and its adjacent genes are mainly enriched in the cell physiology and tumor formation signaling pathways. In brief, all the results summarized the characteristics of NOTCH1–4 mutation status in PCa and indicated that NOTCH1–4 mutations may participate in the development of PCa.
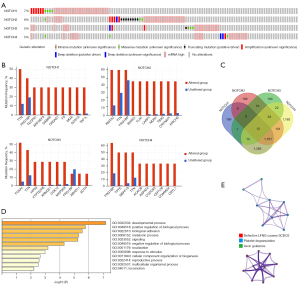
Table 1
| # | Symbol | Description | Category |
|---|---|---|---|
| 1 | TLCD3B | TLC domain containing 3B | Protein coding |
| 2 | DDI1 | DNA damage inducible 1 homolog 1 | Protein coding |
| 3 | CRYL1 | Crystallin lambda 1 | Protein coding |
| 4 | SLC25A23 | Solute carrier family 25 member 23 | Protein coding |
| 5 | TNK2 | Tyrosine kinase non receptor 2 | Protein coding |
| 6 | DDIT4 | DNA damage inducible transcript 4 | Protein coding |
| 7 | NDNF | Neuron derived neurotrophic factor | Protein coding |
| 8 | IL31RA | Interleukin 31 receptor A | Protein coding |
| 9 | TXNDC16 | Thioredoxin domain containing 16 | Protein coding |
| 10 | MLLT1 | MLLT1 super elongation complex Subunit | Protein coding |
| 11 | FGD1 | FYVE, RhoGEF and PH DOMAIN Containing 1 | Protein coding |
| 12 | KIF20A | Kinesin family member 20A | Protein coding |
Table 2
| GO | Category | Description | Count | % | Log10 (P) | Log10 (q) |
|---|---|---|---|---|---|---|
| R-HAS-5083630 | Reactome gene sets | Defective LFNG causes SCDO3 | 4 | 10.81 | –10.89 | –6.54 |
| GO:0002576 | GO biological processes | platelet degranulation | 3 | 8.11 | –3.18 | –0.63 |
| R-HAS-422475 | Reactome gene sets | Axon guidance | 4 | 10.81 | –2.23 | 0.00 |
GO, Gene Ontology.
NOTCH family genes’ mutations and target drug sensitivity
To determine the potential role of antitumor agents, we used the GDSC database to identify potentially sensitive and selective agents for patients with and without mutations of NOTCH1 and NOTCH2 (because of the absence of NOTCH3 and NOTCH4 data in the GDSC database). GDSC screening results showed a large number of candidate drugs targeting NOTCH1–2 mutations. In both individual screenings, the overwhelming majority of NOTCH1 mutation-targeted drugs were -tinib antineoplastic drugs, such as BMS-754807, linsitinib, saracatinib, and erlotinib, which had sensitivity differences between NOTCH1 mutation and wild-type groups, and cells with NOTCH1 mutations were more sensitive to these four drugs than wild-type ones. NOTCH2 mutation seemed to act as an entospletinib desensitizer (Figure 5A). The drug-sensitivity scatterplot is shown in Figure 5B and the details are shown in Table 3. It is worth noting that most of the NOTCH1–2 mutation-related drugs belong to the family of -tinib drugs, which gives us a hint as to how we can focus on the application of -tinib antineoplastic drugs in the treatment of PCa with different NOTCH mutations.
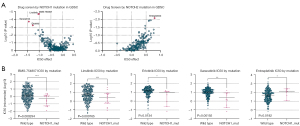
Table 3
| Gene_mutation | Drug | Drug target | Effect size | P value | FDR (%) | Tissue |
|---|---|---|---|---|---|---|
| NOTCH1_mut | BMS-754807 | IGF1R, IR | 0.792 | 0.00817 | 1.16 | PCa |
| NOTCH1_mut | Linsitinib | IGF1R | 0.606 | 0.0112 | 1.35 | PCa |
| NOTCH1_mut | Erlotinib | ABL, SRC | 0.661 | 0.0113 | 5.1 | PCa |
| NOTCH1_mut | Saracatinib | EGFR | 0.593 | 0.0122 | 15.5 | PCa |
| NOTCH2_mut | Entospletinib | SYK | 0.792 | 0.00817 | 43.2 | PCa |
FDR, false discovery rate; Mut, mutation; PCa, prostate cancer.
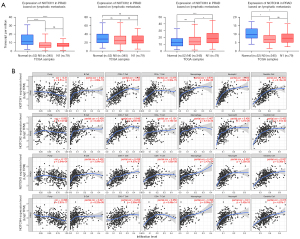
Relationship of NOTCH family genes’ expression to lymph node metastasis and immune cell infiltration in PCa
It has been reported that NOTCH family genes can participate in tissue inflammation and tumor lymphocyte infiltration, and thus may change the tumor’s response to immune-related treatments and the prognosis of patients (30,31). Firstly, we assessed the correlation between NOTCH1–4 expression and lymph node metastasis status. The results indicated that patients with N1 stage exhibited a higher NOTCH3 expression than those with N0 stage (P<0.05). However, there were no significant differences of NOTCH1, NOTCH2 and NOTCH4 expression between patients with N0 and N1 stages (Figure 6A). Furthermore, we used the TIMER database to comprehensively explore the correlation between the expression differences of NOTCH1–4 and immune cell infiltration. As shown in Figure 6B, there was a significant correlation between NOTCH signaling and lymphocyte infiltration, especially CD8+ T cells and CD4+ T cells, which are the key components of tumor immunity. Because it has been reported that there is a significant impact of immune infiltration in the prognosis of various cancer types (32-34), the results indicated that the expression of NOTCH family genes was closely related to the degree of immune infiltration, which then affects the treatment and prognosis in PCa patients.
Discussion
The incidence rate of PCa is one of the highest among the male malignant neoplasms. The clinical manifestations of PCa are heterogeneous. Conventional treatment strategies include reducing the tumor burden and/or testosterone level through radiotherapy, surgery, and/or androgen deprivation. However, PCa inevitably progresses to CRPC, which has limited treatment options and a grim prognosis. It is a matter of the utmost urgency to find new therapeutic targets for PCa. Recently, the presence of lymphocytic infiltrates in PCa tissue suggested an immune mechanism, and thus, targeted immunotherapy is an attractive option for PCa.
This research aimed to expound a joint viewpoint on the potential applications of NOTCH genes as prognostic biomarkers and as molecular drug targets for PCa patients. The tantalizing thing is that NOTCH signaling is paradoxical, being both a tumor inhibitor and promotor (35,36). Correspondingly, our multidimensional analysis found versatility of NOTCH signaling in PCa. Specifically, the transcriptional level of NOTCH1 and NOTCH4 was significantly lower in PCa tissue than in normal tissue. Based on this, NOTCH signaling might be considered a tumor suppressor. However, the analysis from a clinically perspective found that the expression trend of NOTCH3 and NOTCH4 increased as the Gleason score rose. A high expression of NOTCH1, NOTCH3, and NOTCH4 appears to be the cause of the poor prognosis of PCa patients; therefore, the role of NOTCH signaling is much more complicated than simply either being beneficial or harmful in PCa. Two plausible hypotheses may explain this: one, as the main factor in the internal environment, protein plays a pivotal role in homeostasis and functioning. Changes in the transcription level are not a proxy for the physiological process of cells. So, analysis limited to transcription is challenging to depict the overall picture. NOTCH proteins detection should be done in further studies. Another possible reason is that there were more samples in the Gleason score ≤7 groups than in the Gleason score >7 groups. Also, a lower mRNA expression of NOTCH in the Gleason score ≤7 groups may further drag down the overall NOTCH transcription levels; therefore, some further research based on larger samples is required. It is expected that the role of NOTCH family genes in PCa will be better defined with a clearer understanding of the different microenvironments and subtypes with differences in biological characteristics.
Next, we investigated the internal relationship among NOTCH family genes. The moderate-to-high correlations among NOTCH1–4 were evident, and imply a cooperative role of NOTCH family genes working together as a contribution to PCa progression. In addition, it is reported that selective inhibition of NOTCH1 attenuates PCa cell growth in the castrated scenario (37). Some studies have claimed that as a synergistic inhibitor of AR, the NOTCH signaling downstream target gene Hey1 can inhibit the activation of AR and the expression of AR-related genes (27,38), although AR activation could also inhibit NOTCH signaling activation in turn (39). Contrary to previous studies, our data demonstrated that NOTCH1–4 had a positive correlation to AR expression and negatively regulated KLK3 and TMPRSS2. We speculate that the NOTCH family genes perhaps have the ability to prevent AR nuclear translocation and inhibit DNA binding, by which it impedes AR-mediated transcription. The loss of coordination between NOTCH signaling and AR signaling might alter tumor cells’ characteristics. These complicated correlations between the expression of NOTCH family genes and AR/AR-related genes may also result in a higher Gleason score in PCa patients and worse prognosis, and further research still needs to be done.
We also explored if FURIN was the most pertinent interaction factor to NOTCH family genes, and identified a positive correlation between FURIN and NOTCH1–3 at the transcriptional level. FURIN is a widely expressed calcium-dependent protease, and plays a critical role in embryogenesis, as well as catalyzing the maturation of a large number of different pro-protein substrates among which are even some protease systems that regulate diseases. FURIN expression is enhanced in a variety of cancer types, and its activity promotes many cancer-related processes, such as cell proliferation, migration and invasion, and vascularization (40,41). To our knowledge, no study to date has linked up NOTCH signaling and FURIN in PCa. Thus, our results could be the foundation for further investigation.
Finally, current therapies targeting the NOTCH signaling pathway mainly focus on the chemical or biological inhibition of NOTCH family genes (42). However, the development of new drugs has the problems of high cost, long development cycle, and difficulty to adapt to the fast-changing patients’ demand. Here, a new route of drug screening based on NOTCH1–4 mutation status was discussed to broaden the treatment regimen. Despite the missing data for NOTCH3–4 in the GDSC database, the analysis targeting NOTCH1 and NOTCH2 mutations shed light on the role of -tinib antineoplastic drugs in PCa patients. The -tinib antitumor drugs are a new class of biologically targeted cancer drugs, and research has highlighted their role in PCa (43). Our results suggested that the NOTCH genes’ mutation status is a clue for the use of -tinib antitumor drugs in PCa patients. Intriguingly, research has shown that -tinib drugs affect the development of murine PCa by activating antitumor innate immunity (44,45). Consistent with the results above, tumor-infiltrated lymphocyte analysis revealed the relationship between the NOTCH family genes and classical immune cells (B cells, CD8+ T cells, CD4+ T cells, macrophages) infiltration. To sum up, full integration of NOTCH mutation status, -tinib drugs and tumor immune cell infiltration could lead to a bright prospect in the therapeutic management of PCa patients.
Inevitably, there are still some limitations of our study. First, analysis at the transcriptional level alone does not provide a complete landscape of organism function. Another independent cohort study and in vitro or in vivo follow-up may be necessary to verify our results.
Conclusions
We revealed a significant difference in NOTCH family genes’ expression between normal prostate tissue and PCa tissue, and the expression had a positive correlation with Gleason score, DFS, lymph node metastasis, and immune cell infiltration of PCa patients. NOTCH signaling might be involved in tumor promotion and inflammation, and then accelerate tumor progression by inducing AR signaling and FURIN. Patients with NOTCH family gene mutations might be sensitive to -tinib antineoplastic drugs. We hope that our findings give a new perspective on designing new immunotherapy agents, as well as helping clinicians select appropriate drugs and prognostic biomarkers for PCa patients.
Acknowledgments
Funding: This study was supported by the National Natural Science Foundation of China (No. 81802536), and the Fundamental Research Funds for the Central Universities (No. 20ykpy22).
Footnote
Reporting Checklist: The authors have completed the STREGA reporting checklist. Available at https://tau.amegroups.com/article/view/10.21037/tau-22-281/rc
Data Sharing Statement: Available at https://tau.amegroups.com/article/view/10.21037/tau-22-281/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tau.amegroups.com/article/view/10.21037/tau-22-281/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of The Third Affiliated Hospital of Sun Yat-sen University (No. 2019-02-153-01) and informed consent was given by all the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Fuchs HE, et al. Cancer statistics, 2022. CA Cancer J Clin 2022;72:7-33. [Crossref] [PubMed]
- Kan Z, Jaiswal BS, Stinson J, et al. Diverse somatic mutation patterns and pathway alterations in human cancers. Nature 2010;466:869-73. [Crossref] [PubMed]
- Barbieri CE, Baca SC, Lawrence MS, et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat Genet 2012;44:685-9. [Crossref] [PubMed]
- Liu Y, Gusev A, Heng YJ, et al. Somatic mutational profiles and germline polygenic risk scores in human cancer. Genome Med 2022;14:14. [Crossref] [PubMed]
- Blattner M, Lee DJ, O'Reilly C, et al. SPOP mutations in prostate cancer across demographically diverse patient cohorts. Neoplasia 2014;16:14-20. [Crossref] [PubMed]
- Wedge DC, Gundem G, Mitchell T, et al. Sequencing of prostate cancers identifies new cancer genes, routes of progression and drug targets. Nat Genet 2018;50:682-92. [Crossref] [PubMed]
- Su F, Zhang W, Zhang D, et al. Spatial Intratumor Genomic Heterogeneity within Localized Prostate Cancer Revealed by Single-nucleus Sequencing. Eur Urol 2018;74:551-9. [Crossref] [PubMed]
- Heinlein CA, Chang C. Androgen receptor in prostate cancer. Endocr Rev 2004;25:276-308. [Crossref] [PubMed]
- Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science 1999;284:770-6. [Crossref] [PubMed]
- Mumm JS, Schroeter EH, Saxena MT, et al. A ligand-induced extracellular cleavage regulates gamma-secretase-like proteolytic activation of Notch1. Mol Cell 2000;5:197-206. [Crossref] [PubMed]
- Brou C, Logeat F, Gupta N, et al. A novel proteolytic cleavage involved in Notch signaling: the role of the disintegrin-metalloprotease TACE. Mol Cell 2000;5:207-16. [Crossref] [PubMed]
- Aster JC, Pear WS, Blacklow SC. The Varied Roles of Notch in Cancer. Annu Rev Pathol 2017;12:245-75. [Crossref] [PubMed]
- Stopsack KH, Nandakumar S, Wibmer AG, et al. Oncogenic Genomic Alterations, Clinical Phenotypes, and Outcomes in Metastatic Castration-Sensitive Prostate Cancer. Clin Cancer Res 2020;26:3230-8. [Crossref] [PubMed]
- Li JL, Sainson RC, Shi W, et al. Delta-like 4 Notch ligand regulates tumor angiogenesis, improves tumor vascular function, and promotes tumor growth in vivo. Cancer Res 2007;67:11244-53. [Crossref] [PubMed]
- Wang XD, Shou J, Wong P, et al. Notch1-expressing cells are indispensable for prostatic branching morphogenesis during development and re-growth following castration and androgen replacement. J Biol Chem 2004;279:24733-44. [Crossref] [PubMed]
- Shou J, Ross S, Koeppen H, et al. Dynamics of notch expression during murine prostate development and tumorigenesis. Cancer Res 2001;61:7291-7. [PubMed]
- Farah E, Li C, Cheng L, et al. NOTCH signaling is activated in and contributes to resistance in enzalutamide-resistant prostate cancer cells. J Biol Chem 2019;294:8543-54. [Crossref] [PubMed]
- Wang L, Zi H, Luo Y, et al. Inhibition of Notch pathway enhances the anti-tumor effect of docetaxel in prostate cancer stem-like cells. Stem Cell Res Ther 2020;11:258. [Crossref] [PubMed]
- Li T, Fan J, Wang B, et al. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res 2017;77:e108-10. [Crossref] [PubMed]
- Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:pl1. [Crossref] [PubMed]
- Tang Z, Kang B, Li C, et al. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res 2019;47:W556-60. [Crossref] [PubMed]
- Chandrashekar DS, Bashel B, Balasubramanya SAH, et al. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017;19:649-58. [Crossref] [PubMed]
- Warde-Farley D, Donaldson SL, Comes O, et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res 2010;38:W214-20. [Crossref] [PubMed]
- Szklarczyk D, Gable AL, Lyon D, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 2019;47:D607-13. [Crossref] [PubMed]
- Yang W, Soares J, Greninger P, et al. Genomics of Drug Sensitivity in Cancer (GDSC): a resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res 2013;41:D955-61. [Crossref] [PubMed]
- Kamińska A, Pardyak L, Marek S, et al. Notch signaling regulates nuclear androgen receptor AR and membrane androgen receptor ZIP9 in mouse Sertoli cells. Andrology 2020;8:457-72. [Crossref] [PubMed]
- Belandia B, Powell SM, García-Pedrero JM, et al. Hey1, a mediator of notch signaling, is an androgen receptor corepressor. Mol Cell Biol 2005;25:1425-36. [Crossref] [PubMed]
- Thomas G. Furin at the cutting edge: from protein traffic to embryogenesis and disease. Nat Rev Mol Cell Biol 2002;3:753-66. [Crossref] [PubMed]
- Siegfried G, Basak A, Cromlish JA, et al. The secretory proprotein convertases furin, PC5, and PC7 activate VEGF-C to induce tumorigenesis. J Clin Invest 2003;111:1723-32. [Crossref] [PubMed]
- Biktasova AK, Dudimah DF, Uzhachenko RV, et al. Multivalent Forms of the Notch Ligand DLL-1 Enhance Antitumor T-cell Immunity in Lung Cancer and Improve Efficacy of EGFR-Targeted Therapy. Cancer Res 2015;75:4728-41. [Crossref] [PubMed]
- Cheung PF, Neff F, Neander C, et al. Notch-Induced Myeloid Reprogramming in Spontaneous Pancreatic Ductal Adenocarcinoma by Dual Genetic Targeting. Cancer Res 2018;78:4997-5010. [Crossref] [PubMed]
- Sanz-Pamplona R, Melas M, Maoz A, et al. Lymphocytic infiltration in stage II microsatellite stable colorectal tumors: A retrospective prognosis biomarker analysis. PLoS Med 2020;17:e1003292. [Crossref] [PubMed]
- Väyrynen SA, Zhang J, Yuan C, et al. Composition, Spatial Characteristics, and Prognostic Significance of Myeloid Cell Infiltration in Pancreatic Cancer. Clin Cancer Res 2021;27:1069-81. [Crossref] [PubMed]
- Ascierto PA, Lewis KD, Di Giacomo AM, et al. Prognostic impact of baseline tumour immune infiltrate on disease-free survival in patients with completely resected, BRAFv600 mutation-positive melanoma receiving adjuvant vemurafenib. Ann Oncol 2020;31:153-9. [Crossref] [PubMed]
- Guo H, Lu Y, Wang J, et al. Targeting the Notch signaling pathway in cancer therapeutics. Thorac Cancer 2014;5:473-86. [Crossref] [PubMed]
- Deng G, Ma L, Meng Q, et al. Notch signaling in the prostate: critical roles during development and in the hallmarks of prostate cancer biology. J Cancer Res Clin Oncol 2016;142:531-47. [Crossref] [PubMed]
- Cui J, Wang Y, Dong B, et al. Pharmacological inhibition of the Notch pathway enhances the efficacy of androgen deprivation therapy for prostate cancer. Int J Cancer 2018;143:645-56. [Crossref] [PubMed]
- Lavery DN, Villaronga MA, Walker MM, et al. Repression of androgen receptor activity by HEYL, a third member of the Hairy/Enhancer-of-split-related family of Notch effectors. J Biol Chem 2011;286:17796-808. [Crossref] [PubMed]
- Nantermet PV, Xu J, Yu Y, et al. Identification of genetic pathways activated by the androgen receptor during the induction of proliferation in the ventral prostate gland. J Biol Chem 2004;279:1310-22. [Crossref] [PubMed]
- Jaaks P, Bernasconi M. The proprotein convertase furin in tumour progression. Int J Cancer 2017;141:654-63. [Crossref] [PubMed]
- Soylu H, Kırca M, Avcı S, et al. Antiandrogen abiraterone and docetaxel treatments affect Notch1, Jagged1 and Hes1 expressions in metastatic prostate cancer cells. Exp Mol Pathol 2021;119:104607. [Crossref] [PubMed]
- Egloff AM, Grandis JR. Molecular pathways: context-dependent approaches to Notch targeting as cancer therapy. Clin Cancer Res 2012;18:5188-95. [Crossref] [PubMed]
- He Z, Khatib AM, Creemers JWM. The proprotein convertase furin in cancer: more than an oncogene. Oncogene 2022;41:1252-62. [Crossref] [PubMed]
- Patnaik A, Swanson KD, Csizmadia E, et al. Cabozantinib Eradicates Advanced Murine Prostate Cancer by Activating Antitumor Innate Immunity. Cancer Discov 2017;7:750-65. [Crossref] [PubMed]
- Zhou B, Gao S. Pan-Cancer Analysis of FURIN as a Potential Prognostic and Immunological Biomarker. Front Mol Biosci 2021;8:648402. [Crossref] [PubMed]
(English Language Editor: K. Brown)


