
Integrated analysis reveals prognostic value and progression-related role of AMIGO2 in prostate cancer
Introduction
Prostate cancer (PCa) is one of the most frequently diagnosed genitourinary tumor in men, especially in developed countries. Referring to the American Cancer Society of 2021, the number of new cases of prostate cancer diagnosis ranked first in male tumors, and the number of deaths ranked second (1). In European countries, the male incidence rates and mortality rates also come out top. In developing countries, such as China, prostate cancer ranked eighth in the incidence of male cancer, but the incidence is increasing year by year (2,3). By the early state screening of serum prostate specific antigens (PSA) and increasingly improved biopsy methods, the early diagnosis and treatment of prostate cancer is gradually developing in a good direction (4). However, to some extent, variety of current diagnostic and prognostic criterions also led to overdiagnosis and overtreatment (5). Although, numerous translational studies have been conducted to investigate the pathogenesis of prostate cancer and found out several potential biomarkers [e.g., PTEN (6), CD73 (7), BRCA1 (8) and ERG (9)], high level of genetic heterogeneity of PCa and a rigorous and extensive validation is required before we put them in clinical use. Therefore, understanding the cancer-promoting process and its potential molecular mechanism for PCa, enhancing the prognostic abilities and establishing the effective therapeutic strategies were of great essential for improving the survival rate of PCa patients.
The Adhesion Molecule with Ig Like Domain family (AMIGO) was first discovered in the central nervous system and reported to promote neuronal survival by inhibiting the progression of apoptosis, a function associated with social memory formation and mental retardation (10). The AMIGO protein family has three members, AMIGO1, AMIGO2, and AMIGO3, of which AMIGO2 is currently being studied more deeply. AMIGO2 was identified as an interesting tumor-associated gene that was upregulated in most gonadotroph, somatotroph and lactotroph tumors (11). Furthermore, it was reported that AMIGO2 involving in the proliferation and metastasis of several malignant cancer, i.e., melanoma (12). However, the comprehensive analysis related to the diagnostic and prognostic of AMGIO2 in various type of cancer, especially in prostate cancer was not fully recognized.
In present study, we comprehensively analyzed the transcriptome level and prognostic value of AMIGO2 in pan-cancer and illustrated that AMIGO2 was a significant tumor-associated gene in prostate cancer. Integrating analysis of AMGIO2 and various clinical variables by univariate and multivariate Cox regression analysis revealed that AMIGO2 was a reliable prognostic factor for recurrence-free survival (RFS) in prostate cancer. A novel nomogram was constructed to accurately predict 3-, 5- and 7-year of RFS outcome, respectively, for PCa. Furthermore, we identified the central role of AMIGO2 in prostate cancer and that may serve as a progression-related factor via epithelial mesenchymal transition (EMT). We present the following article in accordance with the REMARK reporting checklist (available at https://tau.amegroups.com/article/view/10.21037/tau-21-1148/rc).
Methods
Ethical statement
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Ethical approval was not required because the data used in the present study were obtained from public databases.
Data source and procession
The Cancer Genome Atlas (TCGA) pan-cancer data, including RNA-sequencing data and curated clinical phenotypes of 32 prevalent cancer types, were downloaded from Pan-Cancer Atlas Hub in University of California Santa Cruz (UCSC) Xena dataset (https://xenabrowser.net/). The transcriptome data of AMIGO2 in 32 tumor types, including Adrenocortical carcinoma (ACC, n=77), Bladder Urothelial Carcinoma (BLCA, n=407), Breast invasive carcinoma (BRCA, n=1,091), Cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC, n=304), Cholangiocarcinoma (CHOL, n=36), Colon adenocarcinoma (COAD, n=286), Lymphoid Neoplasm Diffuse Large B-cell Lymphoma (DLBC, n=47), Esophageal carcinoma (ESCA, n=181), Glioblastoma multiforme (GBM, n=152), Head and Neck squamous cell carcinoma (HNSC, n=518), Kidney Chromophobe (KICH, n=66), Kidney renal clear cell carcinoma (KIRC, n=530), Kidney renal papillary cell carcinoma (KIRP, n=288), Brain Lower Grade Glioma (LGG, n=508), Liver hepatocellular carcinoma (LIHC, n=369), Lung adenocarcinoma (LUAD, n=513), Lung squamous cell carcinoma (LUSC, n=498), Mesothelioma (MESO, n=87), Ovarian serous cystadenocarcinoma (OV, n=420), Pancreatic adenocarcinoma (PAAD, n=178), Pheochromocytoma and Paraganglioma (PCPG, n=177), Prostate adenocarcinoma (PRAD, n=495), Rectum adenocarcinoma (READ, n=91), Sarcoma (SARC, n=258), Skin Cutaneous Melanoma (SKCM, n=102), Stomach adenocarcinoma (STAD, n=414), Testicular Germ Cell Tumors(TGCT, n=132), Thyroid carcinoma (THCA, n=504), Thymoma (THYM, n=119), Uterine Corpus Endometrial Carcinoma (UCEC, n=180), Uterine Carcinosarcoma. (UCS, n=57), Uveal Melanoma (UVM, n=79) and relevantly paracancerous tissues were extracted for further analysis. The reads per kilobase per million mapped reads (RPKM) profiling data and corresponding clinical characteristics for DKFZ dataset and Genetic alteration of prostate cancer in TCGA-PRAD cohort were obtained from cBioPortal (https://www.cbioportal.org/) (13,14). Besides, the data of staining profiles for proteins in human tumor tissue based on immunohistochemistry (IHC) using tissue microarrays were downloaded from The Human Protein Atlas (https://www.proteinatlas.org/). DNA methylation data of 495 prostate cancer sample were collected from TCGA-PRAD program. The summary of clinical information of TCGA-PRAD dataset and DKFZ cohort for the patient in this study were shown in Table S1. The National Cancer Institute cell line panel (NCI-60) data, containing compound activity and RNA-seq data from 60 diverse human cancer cell lines, were obtained from UCSC xena (https://xena.ucsc.edu/). Expression data of GSE158494 was obtained from Gene Expression Omnibus (GEO, https://www.ncbi.nlm.nih.gov/geo/).
Pan-cancer differential expression analysis and survival analysis of AMIGO2
Wilcoxon signed-rank test was leveraged to detect the differential expression of AMIGO2 between normal and tumor tissues in pan-cancer. Besides, three or more groups were compared by Kruskal-Wallis test. The prognostic value of AMIGO2 in pan-cancer were determined by Univariate Cox regression analysis and visualized by forest plot. The Kaplan-Meier analysis and the log-rank test were used to compare the RFS outcomes, defined as the length of time a patient survives without any signs or symptoms of cancer after the end of primary cancer treatment, between the high- and low-expression groups based on the median expression level of AMIGO2 using the R package “survival”. All statistical tests were two-sided and P value <0.05 was considered statistically significant.
Cell culture
The prostate cancer cell lines, DU145, PC3, were all recovered from the nitrogen liquid tank in our lab. Cells were cultured in RPMI-1640 medium supplemented with 10% fetal bovine and 1% Penicillin and Streptomycin. All cell lines were maintained at 37 ℃ and 5% CO2.
siRNA transfection and cell proliferation assay
All small interference RNAs (siRNA) were synthesized by GenePharma (Hangzhou, China). DU145 and PC3 cells were seeded into the 6-well plates, then the negative control siRNA and siRNAs targeted to AMIGO2 mixed with siRNA mate (G04003, GenePharma, China) were added to the prepared cells. Western Blotting was used to detect the knockdown efficiency in DU145 and PC3 cells.
Cell proliferation assay
To assess the potential role of AMIGO2 in PCa, we performed cell proliferation assay with Cell Counting Kit-8 (CCK-8, MA0218-500, Meilunbio, China). 2500 transfected cells/well were seeded in 96-well plates, and then used CCK-8 reagent to detect the viability of cells at 6, 24, 48, and 72 h by the absorbance at 450 nm using the microplate reader (Bio-Rad iMark).
Colony formation assay
One thousand cells/well were seeded in 12-well plates and cultured for 7 days. Cells were re-transfected every 72 h. The colonies were finally fixed and stained with 0.1% crystal violet. The result was photographed with camera and the number of colonies were counted.
Western blot analysis
Western blot analysis was performed as our previous report (15). The antibodies were used as followed: AMGIO2 (1:1,000 dilution, sc-373699, SANTA CRUZ, USA), E-Cadherin, N-Cadherin, Vimentin, Slug and Snail (1:1,000 dilution, 49398, CST, USA) and β-Actin (1:5,000 dilution, 3700, CST, USA).
IHC
To further examine the relationship between AMIGO2 and major clinical characteristics of PCa, the protein expression levels of AMIGO2 in PCa were examined by IHC using a PCa tissue microarray (TMA; PR803c, Alenabio, Xi’an, China) cohort according to the protocol of our previous report (16). Detail information of the TMA cohort were provided in Table S1. Rabbit anti-AMIGO2 (sc-373699, SANTA CRUZ, USA) was used at a dilution of 1:50. The AMIGO2 protein level was determined by the percentage and intensity of staining in each sample. The percentage of staining was divided into 4 categories, as follows: 0 (0–5%), 1 (6–25%), 2 (26–50%), 3 (51–75%) and 4 (>75%). The staining intensity was divided into 4 levels, as follows: 0 (negative), 1 (weak), 2 (moderate) and 3 (strong). Then multiplying the staining percentage and staining intensity to get the immunoreactivity score (IRS).
Evaluating the prognostic value of AMIGO2 in prostate cancer
Given the significant role of AMIGO2 in prostate cancer, we further explored the prognostic value of AMIGO2 in PCa. The AMIGO2 and various clinical variables [age, Gleason score (GS) and tumor stage] were analyzed, respectively, using the univariate Cox proportional hazard regression method to explore their relationship with RFS outcome in prostate cancer. The statistically significant predictors (P<0.05) were selected and then common analyzed by multivariate Cox regression analysis. Meanwhile, concordance index (C-index), a metric to evaluate the predictive power, was calculated by function ‘coxph’ in R package ‘survival’. R package ‘rms’ was used to construct a predictive nomogram using the significant prognostic factors for predicting 3-, 5-, and 7-year RFS of PCa patients in the TCGA-PRAD cohort. The calibration curves, created by R package ‘rms’, were used to evaluate the performance of the prognostic nomogram in regard with the predictions for 3-, 5-, or 7-year RFS outcomes.
Exploration of the potentially biological function of AMIGO2 in prostate cancer
In order to explore the potential biological function of AMIGO2 in prostate cancer, two groups, defined by median cut-off value of AMIGO2 (positive and poor outcome of prostate cancer), were used to detect the differential expressed genes (DEGs). Furthermore, Gene Set Enrichment Analysis (GSEA) was further carried out to investigate the significantly DEGs [The Log2|Fold-change| (log2|FC|) >1 and P value <0.05] using the annotation gene sets of ‘Hallmark’ from MsigDB dataset (http://www.broadinstitute.org/gsea) (17,18). False discovery rate (FDR) <0.05, adjusted P value <0.01 and normalized enrichment score (NES) >1 or NES <–1 were considered as statistical significance.
Statistical analysis
Besides the methods aforementioned, other statistical methods used in the study included Kaplan-Meier survival analysis, Receiver operating characteristic curve (ROC) analysis, chi-square test, Pearson’s correlation analysis, and Wilcoxon signed-rank test, with two-tailed P<0.05 being considered as statistical significance level. P value adjustment was applied when multiple comparisons were necessary. All the bioinformatics and statistical analysis were performed in R (version 4.0.5).
Results
Exploration of the expression levels and prognostic value of AMIGO2 in pan-cancer
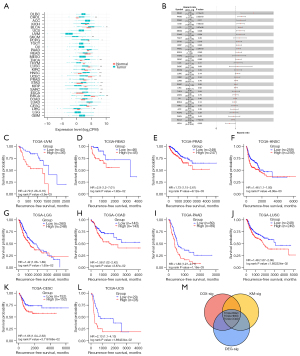
To comprehensive analysis the transcriptome expression pattern of AMIGO2, pan-cancer data, including 33 common cancer type, from TCGA were leveraged. As shown in Figure 1A, AMIGO2 was significantly differentially expressed in multiple cancer type, involving in BLCA, BRCA, CHOL, ESCA, HNSC, KIRC, KIRP, LIHC, LUSC, PRAD, SARC, SKCM, STAD, THCA and UCEC, revealing the significant role of AMIGO2 in cancer occurrence (all P value <0.05). Furthermore, survival analysis was performed to explore the prognostic value of AMIGO2 across cancers. Cox proportional hazards model analysis revealed that AMIGO2 expression levels were associated with relapse-free survival in BLCA, COAD, ESCA, HNSC, KICH, KIRP, LGG, MESO, PAAD, PRAD, READ, STAD and UVM (Figure 1B, all P value <0.05). Kaplan-Meier survival analysis also demonstrated that among patients in CESC, COAD, HNSC, LGG, LUSC, PAAD, PRAD, READ, UCS, UVM, high expression of AMIGO2 had a worse RFS than that in low expression AMIGO2 (Figure 1C-1L). Furthermore, intersection of the significant performance of AMIGO2 in differential expression analysis, Cox regression analysis and Kaplan-Meier analysis, AMIGO2 was showing the significant role in PRAD, HNSC and COAD, which were selected for further analysis (Figure 1M).
Identified the effects of AMIGO2 on the progression of prostate cancer
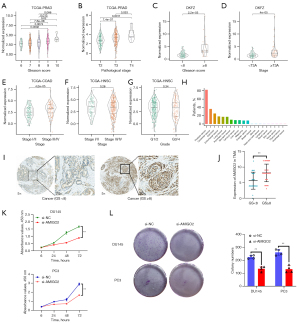
Given the major role of AMIGO2 in PRAD, HNSC and COAD we discovered above, we then explored the expression pattern of AMIGO2 in these three cancers. It is worth to noting that the PCa patients with higher Gleason score (GS) (Figure 2A, P=0.0002, P=0.0074, P=7.6e-06, P=0.02 and P=0.012 for GS =6 vs. GS =9, GS =6 vs. GS =10, GS =7 vs. GS =9, GS =7 vs. GS =10 and GS =8 vs. GS =9, respectively) and worse pathological tumor stage (Figure 2B, P=7.4e-07 for pathological tumor stage =T2 vs. pathological tumor stage =T3 and P=0.0011 for pathological tumor stage =T2 vs. pathological tumor stage =T4), the mRNA expression level of AMIGO2 was significantly increasing in TCGA-PRAD dataset. Moreover, DFKZ dataset was utilized to validated the clinical relevance of AMIGO2 in PCa, the result, consist with above findings, shown the closely relationship between AMIGO2 and PCa progression (Figure 2C,2D, P=2.2e-05 for GS <8 vs. GS ≥8 and P=4e-05 for pathological tumor stage <T3A vs. ≥T3A). Besides, among the COAD patients with worse pathologic stage, AMIGO2 was significantly higher than those with better pathological stage (Figure 2E, P=4.2e-05 for pathological stage I/II vs. stage III/IV). Whereas, there was no significant relationship between AMIGO2 and pathological stage or tumor grade in HNSC (Figure 2F,2G, P>0.05). Moreover, we compared the protein expression levels of AMIGO2 in the Human Protein Atlas dataset. The results suggested that AMIGO2 showed strong granular cytoplasmic positivity in prostate cancer, colorectal cancer, carcinoid, endometrial cancer and lung cancer, of which, AMIGO2 had the highest percentage of the patients with strong staining in prostate cancer, indicating that AMIGO2 might function as an important role in these cancers, especially for the prostate cancer (Figure 2H). Thus, we primarily focused on the potential effect of AMIGO2 on PCa in this study. Then, we further examined the expression pattern of AMIGO2 in prostate cancer tissue. Consist with the result we showed above, AMIGO2 was correlated with the higher GS in prostate cancer (Figure 2I) and statistically significant higher in GS ≥8 compared to GS <8 (Figure 2J). Furthermore, in order to explore the role of AMIGO2 in PCa in vitro, the effect of AMIGO2 in PCa proliferation was detected through growth curves and colony formation in DU145 and PC3. The result shown that reduced expression of AMIGO2 protein inhibited cell proliferation in both DU145 and PC3 cell line (Figure 2K). The colony formation assay also demonstrated that the ability of PCa cell survival while AMIGO2 knockdown was significantly decreased compared with normal control group (Figure 2L).
Identification of AMIGO2 is a reliable prognostic marker for prostate cancer
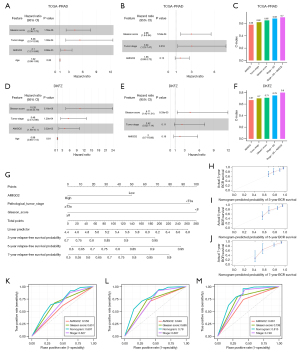
As above mentioned, patient with higher expression of AMIGO2 had worse RFS than that with lower expression of AMIGO2, indicating that AIMGO2 may serve as a prognostic biomarker for predicting the RFS for prostate cancer. To further examined the prognostic value of AMIGO2 in prostate cancer, univariate and multivariate Cox regression analysis were firstly performed in two datasets, i.e., TCGA-PRAD and DFKZ dataset. As shown in Figure 3A, univariate Cox regression analysis revealed that AMIGO2 (HR =2.1, 95% CI: 1.21 to 3.66, P=8.68e-03), GS (HR =4.27, 95% CI: 2.36 to 7.72, P=1.55e-06) and tumor stage (HR =5.38, 95% CI: 2.3 to 12.58, P=1.05e-04) were correlated with the relapse-free survival of prostate cancer. Then, these three statistically significant factors (AMIGO2, GS, tumor stage) were further fitting into the multivariate cox regression analysis. The result shown that GS (P=5.54e-04) and tumor stage (P=0.016) were the independent prognostic factor of prostate cancer (Figure 3B). Although AMIGO2 did not show the independent prognostic value of prostate cancer, the C-index of AMIGO2 which combined with Gleason score and tumor stage (C-index: 0.7) was higher than traditional prognostic evaluation system, e.g., T stage and Gleason score, even higher than the C-index of combination with these two clinically prognostic factors (C-index for stage: 0.62, C-index for GS: 0.65 and C-index for combining stage and GS: 0.68, Figure 3C), indicating that AMIGO2 was a reliable prognostic marker providing additional information that supplement the currently used prognosis evaluation system (e.g., T stage and GS), yielding improved predicting models for the prognosis of prostate cancer. Furthermore, we validated the prognostic value of AMIGO2 in an independent cohort (DKFZ). In line with the result in TCGA-PRAD, AMIGO2 shown the prognostic value (HR =4, 95% CI: 1.59 to 10.1, P=3.32e-03; Figure 3D) in prostate cancer and the valuable function of prognostic supplement (Figure 3E,3F). Given the significant prognostic value of AMIGO2 and its relationship with tumor stage and GS in prostate cancer, we developed nomograms for predicting patients’ 3-, 5- and 7-year RFS outcomes, respectively (Figure 3G). The calibration curves in Figure 3H-3J indicated that the prediction performances of these nomograms are close to ideal model (diagonal line). Figure 3K-3M shown that the area under curve (AUC) of the nomograms were 0.697, 0.78 and 0.815 for 3-, 5- and 7-year RFS, respectively. These AUC values of the nomograms were greater than those of the single predictor (i.e., AMIGO2, tumor stage and GS), indicating an advantage of combining these risk factors for PCa prognosis.
Analysis of the potential genetic and epigenetic alterations associated with AMIGO2 in prostate cancer
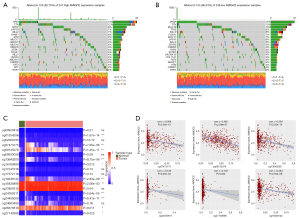
Genomic differences in somatic mutation under different levels of AMIGO2 were further investigated based on the median value of transcriptome expression. As shown in Figure 4A,4B, we displayed the top 20 genomic differences of somatic mutation in high and low expression of AMIGO2, respectively. The result revealed that genomic mutation altered in 155 (62.75%) of 247 AMIGO2 high expression patients and mutation alteration occurred in 165 (66.53%) of 248 AMIGO2 low expression patients. Furthermore, the most common mutation alteration in AMIGO2 high expression group is TP53 (13%, 33/247) and SPOP (15%, 37/248). In addition, TTN, MLL2, MUC17, SPTA1 were the common mutation among top10 mutated genes in both two groups, whereas, LRP1B was more frequently mutated in AMIGO2 high expression group (7%, 18/247) than AMIGO2 low expression group (2%, 6/248). It is noted that Wang et al. (19) had demonstrated down-regulation of LRP1B might promote the migration of colon cancer cells by inducing EMT. Thus, the result of copy number variations (CNV) analysis highlighted the closely relationship between AMIGO2 and EMT. In addition to genetic alterations, analysis integrating transcriptome and methylation been considered as a useful way to explore the fundamental molecular function of AMIGO2 in prostate cancer. Thus, in order to investigate the significant role of AMIGO2 in prostate cancer at CpG methylation level, we explored 18 CpG sites of 50 normal tissue and 502 tumor tissues from TCGA-PRAD cohort. Eleven out of 18 CpG sites (cg01354296, cg07473175, cg08135379, cg13640200, cg16780890, cg18348142, cg18436898, cg24050511, cg24458009, cg26378518, cg27149388) were differentially expressed between normal and tumor tissues (Figure 4C, Wilcoxon signed-rank test, all P<0.05). Furthermore, Pearson correlation analysis indicated that cg07473175, cg08135379, cg13640200, cg24050511, cg27149388, cg18348142, among the 11 differentially expressed CpG, were negatively correlation with AMIGO2 expression (Figure 4D). Taken together, these data suggested that genetic and epigenetic Alterations regulated the abnormal expression of AMIGO2 in PRAD. More importantly, the mutation analysis suggested the closely relationship between AMIGO2 and EMT, and further research about this relation is also needed.
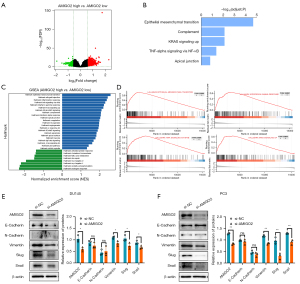
High expression levels of AMIGO2 is correlated with inducing EMT
In order to further explore the potentially biological function of AMIGO2 in prostate cancer, we examined the DEGs between AMIGO2 high and AMIGO2 low expression patients based on the median cut-off value. The volcano plot shown 75 genes were differentially expressed in these two groups (Figure 5A, |log2FC|>1.5, FDR <0.05). Next, biological pathway enrichment analysis was performed using the DEGs. The result demonstrated that genes differentially expressed between AMIGO2 high and low expression group were enriched in cancer-related pathway (i.e., EMT, KRAS signaling, TNF-alpha via NF-KB signaling) and immune-related pathway (complement), indicating that AMIGO2 was significantly correlated with cancer-related pathway and immune-related pathway (Figure 5B). For further validated the central role of AMIGO2 in prostate cancer, AMIGO2 related pathway were analyzed by GSEA. The significant pathways were displayed in Figure 5C. in line with the founding we discovered above, EMT, KRAS signaling, TNF-alpha via NF-KB signaling and immune respond pathway (interferon gamma response) were involving in AMIGO2 high expression group (Figure 5D; NES >1, adjust P<0.01). These results indicated that AMIGO2-induced tumor cell metastasis may be associated with the regulation of EMT progression. To confirm the hypothesis, we examined the expression of EMT-related marker while down-regulated the expression of AMIGO2. As shown in Figure 5E,5F, si-AMIGO2 decreased the expression of Vimentin, Slug and Snail compared with normal control in DU145 and PC3.
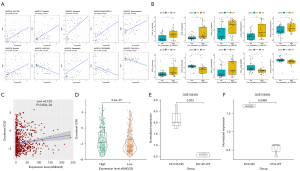
Differential expression levels of AMIGO2 were relevance with docetaxel resistance
In order to explore the relationship between differential expression levels of AMIGO2 and drug treatment response, the relationship between drug sensitivity for 349 anti-cancer compounds and expression of AMIGO2 were detected by Pearson correlation analysis and Wilcoxon signed-rank test. The result shown that AMIGO2 was significantly correlated with 33 out of 349 anti-cancer compounds (P<0.01, Table S2). The top 10 significantly correlated anti-cancer compounds and corresponding boxplot were showing in Figure 6A,6B, and we found that AMIGO2 was positively strong correlation with the compound activity of XAV-939, Sapitinib, Dasatinib, BMS-690514, Staurosporine, Ibrutinib, Saracatinib [correlation coefficient (cor) >0.4 and P<0.001] and negative correlation with ONX-0914, SB-590885, AMG-900 (cor <−0.4 and P<0.001). Besides, cell line with higher expression levels of AMIGO2 displayed higher compound activity of XAV-939, Sapitinib, Dasatinib, BMS-690514, Staurosporine, Ibrutinib, Saracatinib and lower IC50 of ONX-0914, SB-590885, AMG-900 (Wilcoxon signed-rank test, all P<0.05), which indicated the important role of AMIGO2 in anti-cancer drug resistance. Docetaxel is an important antitumor drug for prostate cancer, and its drug resistance problem has always plagued us. Therefore, Given the high correlation between AMIGO2 and the half maximal inhibitory concentration (IC50) of 33 anti-cancer compounds, we further explored the relationship between AMIGO2 and docetaxel resistance. Similarly, the expression levels of AMIGO2 shown positively high correlation with the IC50 of docetaxel (cor =0.126 and P=3.62e-04, Figure 6C) and higher expression of AMIGO2 revealed higher IC50 of docetaxel (Wilcoxon signed-rank test, P=5.2e-07, Figure 6D). Furthermore, an independent dataset, which contained the whole-genome arrays of docetaxel-resistant cells (DU145-DR and PC3-DR) and corresponding wild-type cells (DU145 and PC3), was utilized to verified the effect of AMIGO2 in docetaxel resistance. We found that AMIGO2 was significantly higher expression in DU145-DR (P=0.033) and PC3-DR (P=0.0069) than those in wild-type cells (Figure 6E,6F), revealing that AMIGO2 overexpression promotes docetaxel-resistance in prostate cancer.
Discussion
Globally, prostate cancer is currently the second most common incidence among men. At the same time, prostate cancer has obvious clinical multifocal and polymorphism (20). A variety of potential pathogenesis, such as epithelial cell gene mutations, inflammatory cell infiltration and effects, tumor-related microenvironmental changes, etc., can affect prostate cancer differentiation, phenotype, clinical progression, and metastasis (19). Among them, the influence of tumor microenvironment on tumor epithelial cells has been extensively studied recently, including immune microenvironment and regulation and stromal cell-epithelial cell interaction (21).
In the present study, we investigated the expression levels and prognostic value of AMIGO2 in pan-cancer. The result shown that AMIGO2 was differentially expressed between tumor tissue and normal tissue in multiple cancer type, including BLCA, BRCA, CHOL, ESCA, HNSC, KIRC, KIRP, LIHC, LUSC, PRAD, SARC, SKCM, STAD, THCA, UCEC, indicating the diagnostic value and tumorigenic function of AMIGO2 in diverse cancer type. Nevertheless, AMIGO2 was a promising prognostic factor in different cancer type, i.e., CESC, COAD, HNSC, LGG, LUSC, PAAD, PRAD, READ, UCS, UVM, showing the significant role of AMIGO2 in the occurrence and development of tumors. Emerging study have mentioned that the potentially biological function of AMIGO2 in cancer. It was noticed that patients with high mRNA expression of AMIGO2 experienced significantly shorter survival, suggested that AMIGO2 may serve as a prognostic biomarker for gastric cancer (22). In malignant tumors, Kanda et al. injected a QRsP-11 fibrosarcoma cell line into the mouse spleen and obtained a liver metastasis subgroup LV12, and found that the level of AMIGO2 in LV12 was increased and promoted the attachment/metastasis of tumor cells to hepatocytes (23). In addition to the aforementioned studies, other studies also have shown that AMIGO2 can significantly affect immune T cell immunity. Li et al. demonstrated that AMIGO2 is important in regulating T-cell functions, and may be harnessed as a potential therapeutic target for multiple sclerosis (24). Consistent with the result we shown above, high expression of AMIGO2 was correlated with the immune respond in prostate cancer, indicated that AMIGO2 may serve as an immune-therapy target for prostate cancer. However, the function of AMIGO2 in immune still need to further research. Besides, we identified that AMGIO2 as a prognostic biomarker of prostate cancer that contributing to the progression and aggressive malignant phenotype by regulating EMT in prostate cancer. The initiation of EMT is considered the first step promoting cancer progression that is expected to contribute to the poor prognosis of cancer patient. Therefore, targeting EMT may improve the overall survival rate of patients (25). By using comprehensive bioinformatic analysis, i.e., correlation analysis, biological pathway analysis, Genetic and Epigenetic Alterations displayed the relation between AMIGO2 and EMT. Moreover, two PCa cell lines (PC3 and DU145) were examined that down-regulation of AMIGO2 induced the suppression of EMT-related markers, such as Vimentin, Slug and Snail. Vimentin is a cytoskeletal protein which up-regulation was demonstrated to be involved with poorer outcomes in multiple cancers such breast, gastrointestinal, and prostate cancers (26-28). Vimentin involves in regulating cell migration, cell adhesion, EMT signaling pathways and cytoskeletal reorganization by regulating EMT (29). Besides, Slug and snail are core EMT transcription factors to serve central roles in the execution of EMT in many kinds of biological pathway, e.g., controlling cell–cell adhesion, cell migration and extracellular matrix (ECM) degradation (30). We demonstrated that AMIGO2 promotes PCa progression by altering EMT-related biomarkers and AMIGO2 may be a potential target for PCa therapy. However, the molecular mechanism of the cancer-promoting effect of AMIGO2 in PCa has not yet been fully discovered, and further research is urgently needed. Importantly, we explored the relationship between AMIGO2 and docetaxel resistance and found that AMIGO2 mediates docetaxel resistance in prostate cancer and novel strategy for targeting this gene could provide clinical insights into chemoresistance of prostate cancer.
Conclusions
In conclusion, our first pan-cancer analyses of AMIGO2 demonstrated the significant relationship of AMGIO2 expression with oncogenic and prognostic role in multiple cancer type. Further analysis revealed that AMIGO2 served as a prognostic biomarker in PCa and supplied the external accuracy for predicting RFS in PCa while combined with traditional prognostic evaluation system. Our comprehensive bioinformatics analysis and in vitro examination also suggested that AMIGO2 might play an important role in the progression of PCa, serving as a tumor promoter via EMT. Furthermore, the correlation between AMIGO2 and docetaxel resistance in prostate cancer could provide clinical insights into chemoresistance of prostate cancer.
Acknowledgments
Funding: This work was supported by grants from the National Natural Science Foundation of China (No. 82072813 to WZ); Supporting funding of Guangzhou First People’s Hospital (No. PT81600620 to ZH); Natural Science Foundation of Guangdong Province (Nos. 2020A1515010473, 2018A030313668, and 2020A1515110792, to ZH, YL and JY); China Postdoctoral Science Foundation (No. 2020M682666 to JY).
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at https://tau.amegroups.com/article/view/10.21037/tau-21-1148/rc
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tau.amegroups.com/article/view/10.21037/tau-21-1148/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Ethical approval was not required because the data used in the present study were obtained from public databases.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Fuchs HE, et al. Cancer Statistics, 2021. CA Cancer J Clin 2021;71:7-33. [Crossref] [PubMed]
- Baade PD, Youlden DR, Cramb SM, et al. Epidemiology of prostate cancer in the Asia-Pacific region. Prostate Int 2013;1:47-58. [Crossref] [PubMed]
- Feng RM, Zong YN, Cao SM, et al. Current cancer situation in China: good or bad news from the 2018 Global Cancer Statistics? Cancer Commun (Lond) 2019;39:22. [Crossref] [PubMed]
- Chang AJ, Autio KA, Roach M 3rd, et al. High-risk prostate cancer-classification and therapy. Nat Rev Clin Oncol 2014;11:308-23. [Crossref] [PubMed]
- Bangma CH, Roemeling S, Schröder FH. Overdiagnosis and overtreatment of early detected prostate cancer. World J Urol 2007;25:3-9. [Crossref] [PubMed]
- Boutros PC, Fraser M, Harding NJ, et al. Spatial genomic heterogeneity within localized, multifocal prostate cancer. Nat Genet 2015;47:736-45. [Crossref] [PubMed]
- Leclerc BG, Charlebois R, Chouinard G, et al. CD73 Expression Is an Independent Prognostic Factor in Prostate Cancer. Clin Cancer Res 2016;22:158-66. [Crossref] [PubMed]
- Castro E, Goh C, Olmos D, et al. Germline BRCA mutations are associated with higher risk of nodal involvement, distant metastasis, and poor survival outcomes in prostate cancer. J Clin Oncol 2013;31:1748-57. [Crossref] [PubMed]
- Lokman U, Erickson AM, Vasarainen H, et al. PTEN Loss but Not ERG Expression in Diagnostic Biopsies Is Associated with Increased Risk of Progression and Adverse Surgical Findings in Men with Prostate Cancer on Active Surveillance. Eur Urol Focus 2018;4:867-73. [Crossref] [PubMed]
- Kuja-Panula J, Kiiltomäki M, Yamashiro T, et al. AMIGO, a transmembrane protein implicated in axon tract development, defines a novel protein family with leucine-rich repeats. J Cell Biol 2003;160:963-73. [Crossref] [PubMed]
- Cui Y, Li C, Jiang Z, et al. Single-cell transcriptome and genome analyses of pituitary neuroendocrine tumors. Neuro Oncol 2021;23:1859-71. [Crossref] [PubMed]
- Fontanals-Cirera B, Hasson D, Vardabasso C, et al. Harnessing BET Inhibitor Sensitivity Reveals AMIGO2 as a Melanoma Survival Gene. Mol Cell 2017;68:731-44.e9. [Crossref] [PubMed]
- Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:pl1. [Crossref] [PubMed]
- Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012;2:401-4. [Crossref] [PubMed]
- Xie J, Ye J, Cai Z, et al. GPD1 Enhances the Anticancer Effects of Metformin by Synergistically Increasing Total Cellular Glycerol-3-Phosphate. Cancer Res 2020;80:2150-62. [Crossref] [PubMed]
- Cai Z, Deng Y, Ye J, et al. Aberrant Expression of Citrate Synthase is Linked to Disease Progression and Clinical Outcome in Prostate Cancer. Cancer Manag Res 2020;12:6149-63. [Crossref] [PubMed]
- Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545-50. [Crossref] [PubMed]
- Mootha VK, Lindgren CM, Eriksson KF, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet 2003;34:267-73. [Crossref] [PubMed]
- Haffner MC, Zwart W, Roudier MP, et al. Genomic and phenotypic heterogeneity in prostate cancer. Nat Rev Urol 2021;18:79-92. [Crossref] [PubMed]
- Wang G, Zhao D, Spring DJ, et al. Genetics and biology of prostate cancer. Genes Dev 2018;32:1105-40. [Crossref] [PubMed]
- Condon MS. The role of the stromal microenvironment in prostate cancer. Semin Cancer Biol 2005;15:132-7. [Crossref] [PubMed]
- Nakamura S, Kanda M, Shimizu D, et al. AMIGO2 Expression as a Potential Prognostic Biomarker for Gastric Cancer. Anticancer Res 2020;40:6713-21. [Crossref] [PubMed]
- Kanda Y, Osaki M, Onuma K, et al. Amigo2-upregulation in Tumour Cells Facilitates Their Attachment to Liver Endothelial Cells Resulting in Liver Metastases. Sci Rep 2017;7:43567. [Crossref] [PubMed]
- Li Z, Khan MM, Kuja-Panula J, et al. AMIGO2 modulates T cell functions and its deficiency in mice ameliorates experimental autoimmune encephalomyelitis. Brain Behav Immun 2017;62:110-23. [Crossref] [PubMed]
- Lo UG, Lee CF, Lee MS, et al. The Role and Mechanism of Epithelial-to-Mesenchymal Transition in Prostate Cancer Progression. Int J Mol Sci 2017;18. [Crossref] [PubMed]
- Zhu QS, Rosenblatt K, Huang KL, et al. Vimentin is a novel AKT1 target mediating motility and invasion. Oncogene 2011;30:457-70. [Crossref] [PubMed]
- Kidd ME, Shumaker DK, Ridge KM. The role of vimentin intermediate filaments in the progression of lung cancer. Am J Respir Cell Mol Biol 2014;50:1-6. [Crossref] [PubMed]
- Satelli A, Li S. Vimentin in cancer and its potential as a molecular target for cancer therapy. Cell Mol Life Sci 2011;68:3033-46. [Crossref] [PubMed]
- Herrmann H, Fouquet B, Franke WW. Expression of intermediate filament proteins during development of Xenopus laevis. I. cDNA clones encoding different forms of vimentin. Development 1989;105:279-98. [Crossref] [PubMed]
- Yang J, Antin P, Berx G, et al. Guidelines and definitions for research on epithelial-mesenchymal transition. Nat Rev Mol Cell Biol 2020;21:341-52. [Crossref] [PubMed]


