
Identification of a novel peripheral blood signature diagnosing subclinical acute rejection after renal transplantation
Introduction
Subclinical acute rejection (subAR) is defined by histologically acute rejection without renal dysfunction and correlated with poor graft outcomes, including subsequent acute rejection, de novo donor-specific antibody (dnDSA) production, and graft fibrosis (1-5). The subAR occurs in 29% and 17% of patients with kidney transplantation individually in the 1–2 and 2–3 months (1). Although true randomized, prospective large trials are still lacking, several studies indicated that treatment of subAR was beneficial and led to improved short (6) and long-term renal function (5,7). Therefore, early diagnosis and treatment of subAR is particularly important for preserving renal function and improving the consequences of kidney recipients beyond the early period.
At present, the gold standard for subAR diagnosis remains invasive surveillance biopsies. It is one method to detect subAR patients with stable renal function by performing routine protocol biopsies (1). However, it is constrained by infection risk, sampling error, assessment variability, and other factors (8). To avoid these issues, some studies have tested non-invasive profiles of urinary proteins and blood transcriptomic signatures, but the results have been inconsistent so far (2,9-13). Therefore, there is a clear need for the development of a non-invasive, simple, and accurate biomarker for subAR diagnosis.
The support vector machine recursive feature elimination (SVM-RFE), a feature elimination strategy, is used to screen differentially important features along with high classification accuracies compared with the SVM-RFE (14). In this study, we applied SVM-RFE, least absolute shrinkage and selection operator (LASSO) logistic regression to identify candidate genes for subAR diagnosis in peripheral blood RNA-sequencing (RNA-seq) and microarray datasets of subAR patients from the Gene Expression Omnibus (GEO) database. Then, the stepwise logistic regression method was utilized to construct a diagnostic model that might be potentially implemented in clinical application. Although the fact that prior research has established gene signatures for subAR diagnosis, these signatures were constituted of dozens of genes, making them difficult to translate into clinical practice, and their diagnostic power was significantly reduced in other independent datasets (2,3). Thus, we sought to develop a new peripheral diagnostic model of subAR with an optimal number of genes and reliable performance. The diagnostic model may prove to be a useful tool in clinical practice in the future. Furthermore, we used gene set enrichment analysis (GSEA) and immune cell analysis to investigate the mechanisms underlying the disease in subAR patients. We present the following article in accordance with the TRIPOD reporting checklist (available at https://tau.amegroups.com/article/view/10.21037/tau-22-266/rc).
Methods
The collection and pretreatment of data
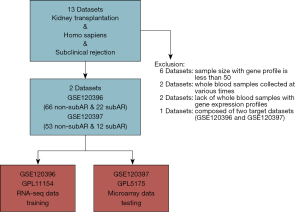
Datasets that met the criteria were included: (I) the size of samples for gene expression profiles is greater than fifty; (II) whole blood specimens with genomic data and biopsy-proven graft status; (III) whole blood samples collected at the same time; (IV) focusing on subAR. Then, two cohorts with gene expression profiles of whole blood cells derived from matched renal transplant patients with unambiguous biopsy-proven subAR or non-subAR were utilized for deeper investigation (Figure 1). The GSE120396 (n=88) RNA-seq dataset contained 66 non-subAR and 22 subAR specimens, and the GSE120397 (n=65) microarray dataset contained 53 non-subAR and 12 subAR specimens (3). The gene expression profiles were normalized using the “limma” R package (15).
GEO data access regulations were strictly followed during data collection and preparation. All analyses were carried out under applicable laws and regulations.
The study was conducted in according to the Declaration of Helsinki (as revised in 2013).
Study design
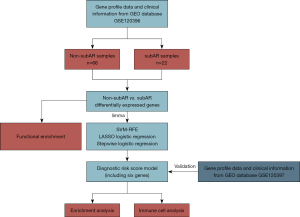
The process of our study was illustrated in Figure 2. In the data collection stage, the GSE120396 (n=88) RNA-seq dataset and the GSE120397 (n=65) microarray dataset were screened. GSE120396 was utilized to identify differentially expressed genes (DEGs) with the threshold of P<0.005 using the “limma” package, followed by SVM-RFE and LASSO logistic regression to select candidate biomarkers for the diagnostic model. The stepwise logistic regression method was applied to screen candidate biomarkers and build a diagnostic signature with low heterogeneity and high consistency. Besides, the performance of the model was validated with the GSE120397 microarray dataset. GSEA and immune-cell analysis were applied in the GSE120396 and GSE120397 datasets to acquire a robust correlation between the model, pathway analysis, and immune-cell analysis in kidney transplant samples.
Gene selection process
The SVM-RFE, a feature elimination strategy, is used to screen differentially important features along with high classification accuracies compared with the SVM-RFE. To list the ranking weights for all genes and arrange genes according to their weight vectors, the SVM-RFE and t-statistics are used in this algorithm. The weights of genes are calculated using the difference between compared groups, which is represented by the expression. After the iteration process, the functionality is removed backward. The dataset is iterated until only one characteristic remain. This study uses the R package “sigFeature” for SVM-RFE (14). The LASSO logistic regression was applied to narrow down the list of top-ranked genes using SVM-RFE. During the LASSO logistic regression procedure, the penalty regularization parameter lambda was selected by a ten-fold cross-validated condition using the “glmnet” R package (16). When binomial deviance reached a minimum through the cross-validation, the genes with non-zero coefficients were chosen. These genes were selected for further analysis using the stepwise logistic regression method.
Generation and validation of the six-gene signature
To identify the candidate genes, the stepwise logistic regression method based on the Akaike Information Criterion (AIC) was utilized to build a model. When the lowest AIC was set, the candidate genes were selected and constructed the six-gene signature. The receiver operating characteristic (ROC) curve was engaged to evaluate the diagnostic capacity and the optimal ROC cutoff point (Youden index) was calculated by the “pROC” R package, which corresponds to the point on the ROC curve where the Youden index reached the highest. Besides, external validation was performed using GSE120397 and its optimal ROC cutoff point. All patients were categorized into high-risk and low-risk groups with their optimal cutoff values respectively.
Function enrichment analysis
Metascape (https://metascape.org/gp/index.html#/main/step1) is an analytical tool for gene function that integrates reputable data repositories (17). Metascape was employed to investigate the possible signaling pathways involved in the subAR using DEGs screened between subAR and non-subAR patients. At least three genes must overlap and enrichment scores of at least 1.5 were required to meet the cutoff, which was determined by a P value cutoff of adjusted 0.01. At least three genes must overlap and enrichment scores of at least 1.5 were required to meet the threshold (adjusted P value <0.01).
GSEA and evaluation of immune cells
By utilizing the “clusterProfiler” R package and a gene set file annotated with the quotation gene set file (c5.go.bp.v7.4.symbols.gmt) we were able to identify biological processes that may be more prevalent in the high-risk patients (18).
The single-sample GSEA with the “GSVA” R package was used to compute the amounts of 28 immune cells (19). Twenty-eight immune cells from two types of immunity (innate and adaptive) were enrolled to explore the relationship between levels of immune cells and risk scores calculated by the diagnostic model.
Statistical analysis
The Pearson correlation test was conducted when the data passed the normality test. The Spearman correlation test was applied for the immune cell analysis. Statistical significance was established at the five percent crucial point (P<0.05) for all data, with the exception of DEG analysis (P<0.005).
Results
The collection and pretreatment of data and study design
We screened the whole blood datasets of subAR from the GEO database to obtain qualified datasets. A more detailed selection process can be detected in the Methods section (Figure 1). There is the flowchart of our study, including training, validation, and further investigation parts (Figure 2). The baseline demographic and clinical characteristics of the subAR and non-subAR groups showed similar graft function at a 3-month surveillance biopsy (3).
Identification of DEGs and functional enrichment analysis
Following the data selection, the genes in GSE120396 and GSE120397 were intersected to obtain the intersection genes (9,750 genes). Then, the intersection genes were performed differential expression analysis between subAR and non-subAR groups in GSE120396. As a consequence, 174 DEGs (55 of which were upregulated and 119 of which were downregulated) were met the criteria (P<0.05) and detected for further investigation (Figure 3A).
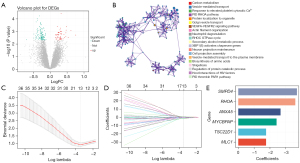
The top 20 biological process clusters, as determined by Metascape, were revealed (Figure 3B). Carbon metabolism, neutrophil degranulation, and other processes were shown to be enriched in the DEGs between subAR and non-subAR patients.
Construction and validation of the diagnostic model
To recognize the subAR-specific expression patterns of genes, SVM-RFE was adopted. Top-ranked 50 DEGs were appropriate for further analysis. After further selecting 50 DEGs with LASSO logistic regression, thirteen biomarkers were selected as the candidate biomarkers of the diagnostic model (Figure 3C,3D).
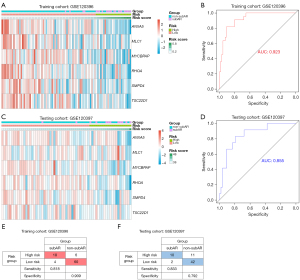
The thirteen candidate genes were input to the stepwise logistic regression method to build the diagnostic model. As a result, six genes (ANXA5, SMPD4, RHOA, TSC22D1, MYCBPAP, and MLC1) were used to develop a diagnostic signature (Figure 3E). The following formula was used to determine the risk score: risk score =171.553+ (−2.67148 * expression level of ANXA5) + (−3.70396 * expression level of SMPD4) + (−3.60213 * expression level of RHOA) + (−1.76173 * expression level of TSC22D1) + (−2.42050 * expression level of MYCBPAP) + (−1.75866 * expression level of MLC1). The heatmap expression of above six genes indicated that these genes have subAR-specific expression patterns and ROC curve suggested a high diagnostic performance of the model in the training cohort [area under the curve (AUC) =0.923] (Figure 4A,4B). Patients were divided into high-risk and low-risk groups based on the optimal cutoff value. To demonstrate the diagnostic power of the six-gene signature, we utilized the testing cohort GSE120397. According to a fixed formula with an optimal cutoff value determined from the testing dataset, patients were separated into high-risk and low-risk groups respectively. The heatmap and ROC curve consistently indicated a high degree of reliability in diagnostic power in the testing cohort (AUC =0.855) (Figure 4C,4D). Besides, the tables demonstrated high sensitivity and specificity of training and testing cohorts (Figure 4E,4F). These findings suggested that the power of the diagnostic model was still highly accurate.
GSEA and immune cells analysis
GSEA was applied to identify potential signaling pathways between high-risk and low-risk patients. For high-risk patients, as illustrated in Figure 5A-5D, we observed a significant decreased transcription of genes involved in the metabolic process of reactive oxygen species (ROS) and the regulation of this metabolic process (Figure 5A-5D).
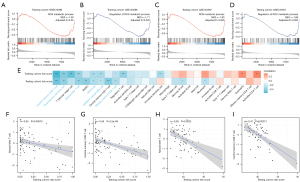
By using single-sample GSEA, we were able to quantify 28 different types of immune cells, including B cells, T cells, monocytes, neutrophils, and others, to better understand the makeup of the peripheral blood. Then, we analyzed the relationship between the risk scores generated by the diagnostic model and immune cells. Our findings revealed that risk scores were negatively related to virtually all innate immune cells and some T cells subtypes (Figure 5E-5I). A consistent trend was observed in the associations of risk scores with immune cells in both the training and testing datasets.
Discussion
SubAR was characterized as biopsies of acute Banff grade 1 or more corresponding with no increased serum creatinine and clinical indications of rejection. Currently, surveillance biopsies were the only method that could diagnose subAR. SubAR might retreat automatically or develop into clinical acute rejection and then lead to the initial onset of chronic rejection (20). Following a thorough screening of the GEO datasets, we selected two datasets focused on subAR, which met the criteria mentioned above. By applying SVM-RFE, LASSO logistic regression, and stepwise logistic regression, we developed a six-gene signature for subAR diagnosis based on publicly available peripheral blood genomic data. The model had high accuracy in both training (AUC =0.923) and validation cohort (AUC =0.855). Once identified in the high-risk group, further tests and treatment were necessary because of the high possibility of subAR detection.
Remarkably, GSEA results indicated that downregulated genes in high-risk patients were associated with the metabolism of ROS and their regulation compared with low-risk patients. Infiltrating monocytes and macrophages themselves forcefully express xanthine oxidoreductase and generate ROS (21). Albrecht et al. showed that infiltrating macrophages seemed to generate ROS and caused the increased level of ROS in the interstitium of chronic kidney transplant failure patients (22). These studies demonstrated the generation of ROS by monocytes and macrophages, suggesting that the reduction in ROS metabolism was related to the decrease of macrophages and monocytes. The immune-cell analysis showed that macrophages and monocytes were negatively correlated with risk scores, supporting that the high-risk patients were with fewer macrophages and monocytes. However, the mechanism needed to be supported by solid evidence from other researches.
Correlation analysis showed that the immune cells (central memory CD8 T cell, Plasmacytoid dendritic cell, natural killer T cell, monocyte, CD56dim natural killer cell, and effector memory CD8 T cell) were statistically significant and consistent in the training and testing cohorts. In contrast to kidney biopsy, which reveals the amount of local immune cell infiltration (23), the peripheral blood transcription profile of kidney transplant patients represents their overall immunological status (24). Immune cell analysis showed that almost all innate immune cells and T cells subtypes, especially for central memory CD8 T cell and natural killer T cell, were reduced in high-risk samples in both training and testing cohorts. Similar phenomena were observed in one study, which revealed the decrease in T cell subtypes in whole blood of patients with kidney rejection (25). These findings indicated that the immunological status in whole blood was complex, and the lack of increased transcription of immune response-related genes may support the hypothesis that immune cells moved from the peripheral blood to kidney allograft (24). Immunological status in whole blood was complex, and the lack of enhanced expression of immune-related genes may support the hypothesis that immune cells traveled from the peripheral blood to the kidney tissue, evidenced by the previous study (24).
Among the six genes in the diagnostic model, the ANXA5 gene encodes the annexin family of calcium-dependent phospholipid-binding proteins, which binds with nanomolar affinity to phosphatidylserines (PS) in a calcium-dependent manner (26,27). One study showed that shielding of exposed PS by the ANXA5 protects against renal ischemia/reperfusion injury and has prognostic significance (28). SMPD4 is a protein-coding gene. SMPD4 is neutral sphingomyelinase with poorly characterized enzymatic activity, which was found mutated in a specific form of congenital microcephaly (29). This gene is activated by DNA damage, cellular stress, and tumor necrosis factor, but it is downregulated by wild-type p53. Among its related pathways are sphingolipid metabolism (REACTOME) and metabolism from the GeneCard database (https://www.genecards.org). RHOA is a protein-coding gene. Among its related pathways is toll-like receptor signaling pathways and DNA damage response (only ATM dependent). This gene encodes a member of the Rho family of small GTPases, which cycle between inactive GDP-bound and active GTP-bound states and function as molecular switches in signal transduction cascades from the GeneCard database (https://www.genecards.org). RHOA was shown to frequently regulate leukocyte-specifically expressed β2 integrins (30) to facilitate phagocytic uptake of pathogens (31), migration (32), and immunological synapse formation (33). TSC22D1, MYCBPAP, and MLC1 are protein-coding genes. The related pathways of TSC22D1 are Development_TGF-beta receptor signaling and ectoderm differentiation. MYCBPAP may play a role in spermatogenesis and be involved in synaptic processes. The related pathways of MLC1 are colorectal cancer metastasis and actin dynamics signaling pathway from the GeneCard database (https://www.genecards.org).
Over the last decades, noninvasive surveillance techniques have become increasingly popular in recent decades for detecting immune-related complications such as subAR. Jackson et al. found that CXCL9 and CXCL10 levels in the urine were significantly increased in recipients with subAR, clinical acute rejection, and BK virus infection but not in stable allograft recipients. Although urine CXCL9 and CXCL10 had higher accuracy of diagnosis compared with serum creatinine, these biomarkers couldn’t distinguish between three diagnoses, including subAR, clinical acute rejection, and BK virus infection (34). Tajima et al. analyzed 80 urinary samples from patients after kidney transplantation and identified human epididymis secretory protein 4 as a diagnostic biomarker (AUC =0.808), which required further verification in independent datasets (12). Friedewald et al. developed a 57-gene biomarker for diagnosing subclinical rejection using peripheral blood samples. However, the excessive number of genes in this model limited its clinical application. In two validation sets, positive predictive values of the 57-gene signature were only 51% and 47% respectively (2). Interestingly, Park et al. innovatively combined blood gene expression and cell-free DNA to diagnose subAR, which performed better than each of assay alone (13). These findings suggested that we may include cell-free DNA as additional input matrices to enrich our gene signature and thus improving the diagnostic performance. Zhang et al. found that a changed diagnostic model composed of seventeen genes could accurately detect the subAR patients from the allograft stable patients. However, the coefficient of each gene in the 17-gene signature constructed in the training cohort was not fixed in the validation cohort, suggesting that genes but not the model were universal in different cohorts (3). While, our study developed a model with only six genes and fixed gene coefficients, which has certain practical potential in clinical utility. The six-gene signature in our study needs prospective studies with a larger sample size to validate its performance in the diagnosis of subAR. Finally, we discussed potential clinical applications of noninvasive biomarkers, such as the period of time for results of testing and costs. The time to library preparation and sequence a sample on specific gene sets takes upwards of a week (35). To our knowledge, the cost comparison between gene expression biomarkers and conventional means (biopsies) is controversial and needed solid evidence to demonstrate. Costs for these techniques (including microarrays and next-generation sequencing technologies) have dropped dramatically over the last decade and are now comparable to other methods utilized routinely in commercial diagnostic laboratories based on improved workflows and analytical tools (36). Puttarajappa et al. raised interesting points that protocol biopsies are more cost-effective methods than noninvasive biomarkers (37). However, there are several concerns about the study by Grewal et al. indicate that analysis by Puttarajappa et al does not provide sufficient evidence to support their conclusion (37,38). Moreover, the gene expression signature for monitoring kidney recipients with stable renal function caused $6,509 savings per year gross versus using surveillance biopsies (38). Furthermore, development of commercial kits for a stable noninvasive biomarker can strengthen the feasibility of testing, shorten the time for library preparation and reduce costs.
Conclusions
In conclusion, a new diagnostic model for subAR with six genes was constructed and verified for subAR patients. Detecting the levels of these six genes may offer a potential tool for the diagnosis of subAR patients. Besides, the model was related to the immunological status and ROS metabolism of subAR patients in whole blood, providing insights into possible potential mechanisms of subAR.
Acknowledgments
Funding: This work was supported by the China Organ Transplantation Development Foundation Elite Program (grant No. YZLC-2021-003), the General Program of the National Natural Science Foundation of China (NSFC) (grant No. 81970645), and the joint fund of Beijing Municipal Commission of Education and Natural Science Foundation of Beijing Municipality (grant No. KZ202010025036).
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://tau.amegroups.com/article/view/10.21037/tau-22-266/rc
Peer Review File: Available at https://tau.amegroups.com/article/view/10.21037/tau-22-266/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tau.amegroups.com/article/view/10.21037/tau-22-266/coif). The authors have no conflicts of interest to declare.
Ethical Statement:
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Nankivell BJ, Chapman JR. The significance of subclinical rejection and the value of protocol biopsies. Am J Transplant 2006;6:2006-12. [Crossref] [PubMed]
- Friedewald JJ, Kurian SM, Heilman RL, et al. Development and clinical validity of a novel blood-based molecular biomarker for subclinical acute rejection following kidney transplant. Am J Transplant 2019;19:98-109. [Crossref] [PubMed]
- Zhang W, Yi Z, Keung KL, et al. A Peripheral Blood Gene Expression Signature to Diagnose Subclinical Acute Rejection. J Am Soc Nephrol 2019;30:1481-94. [Crossref] [PubMed]
- Seifert ME, Yanik MV, Feig DI, et al. Subclinical inflammation phenotypes and long-term outcomes after pediatric kidney transplantation. Am J Transplant 2018;18:2189-99. [Crossref] [PubMed]
- Seifert ME, Agarwal G, Bernard M, et al. Impact of Subclinical Borderline Inflammation on Kidney Transplant Outcomes. Transplant Direct 2021;7:e663. [Crossref] [PubMed]
- Kurtkoti J, Sakhuja V, Sud K, et al. The utility of 1- and 3-month protocol biopsies on renal allograft function: a randomized controlled study. Am J Transplant 2008;8:317-23. [Crossref] [PubMed]
- Loupy A, Vernerey D, Tinel C, et al. Subclinical Rejection Phenotypes at 1 Year Post-Transplant and Outcome of Kidney Allografts. J Am Soc Nephrol 2015;26:1721-31. [Crossref] [PubMed]
- Morgan TA, Chandran S, Burger IM, et al. Complications of Ultrasound-Guided Renal Transplant Biopsies. Am J Transplant 2016;16:1298-305. [Crossref] [PubMed]
- Hricik DE, Nickerson P, Formica RN, et al. Multicenter validation of urinary CXCL9 as a risk-stratifying biomarker for kidney transplant injury. Am J Transplant 2013;13:2634-44. [Crossref] [PubMed]
- Suthanthiran M, Schwartz JE, Ding R, et al. Urinary-cell mRNA profile and acute cellular rejection in kidney allografts. N Engl J Med 2013;369:20-31. [Crossref] [PubMed]
- Roedder S, Sigdel T, Salomonis N, et al. The kSORT assay to detect renal transplant patients at high risk for acute rejection: results of the multicenter AART study. PLoS Med 2014;11:e1001759. [Crossref] [PubMed]
- Tajima S, Fu R, Shigematsu T, et al. Urinary Human Epididymis Secretory Protein 4 as a Useful Biomarker for Subclinical Acute Rejection Three Months after Kidney Transplantation. Int J Mol Sci 2019;20:4699. [Crossref] [PubMed]
- Park S, Guo K, Heilman RL, et al. Combining Blood Gene Expression and Cellfree DNA to Diagnose Subclinical Rejection in Kidney Transplant Recipients. Clin J Am Soc Nephrol 2021;16:1539-51. [Crossref] [PubMed]
- Das P, Roychowdhury A, Das S, et al. sigFeature: Novel Significant Feature Selection Method for Classification of Gene Expression Data Using Support Vector Machine and t Statistic. Front Genet 2020;11:247. [Crossref] [PubMed]
- Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43:e47. [Crossref] [PubMed]
- Friedman J, Hastie T, Tibshirani R. Regularization Paths for Generalized Linear Models via Coordinate Descent. J Stat Softw 2010;33:1-22. [Crossref] [PubMed]
- Zhou Y, Zhou B, Pache L, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun 2019;10:1523. [Crossref] [PubMed]
- Yu G, Wang LG, Han Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 2012;16:284-7. [Crossref] [PubMed]
- Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics 2013;14:7. [Crossref] [PubMed]
- Kumar MS, Heifets M, Moritz MJ, et al. Safety and efficacy of steroid withdrawal two days after kidney transplantation: analysis of results at three years. Transplantation 2006;81:832-9. [Crossref] [PubMed]
- Sun K, Kiss E, Bedke J, et al. Role of xanthine oxidoreductase in experimental acute renal-allograft rejection. Transplantation 2004;77:1683-92. [Crossref] [PubMed]
- Albrecht EW, Stegeman CA, Tiebosch AT, et al. Expression of inducible and endothelial nitric oxide synthases, formation of peroxynitrite and reactive oxygen species in human chronic renal transplant failure. Am J Transplant 2002;2:448-53. [Crossref] [PubMed]
- Sarwal M, Chua MS, Kambham N, et al. Molecular heterogeneity in acute renal allograft rejection identified by DNA microarray profiling. N Engl J Med 2003;349:125-38. [Crossref] [PubMed]
- Viklicky O, Novotny M, Hruba P. Future developments in kidney transplantation. Curr Opin Organ Transplant 2020;25:92-8. [Crossref] [PubMed]
- Mirzakhani M, Shahbazi M, Oliaei F, et al. Immunological biomarkers of tolerance in human kidney transplantation: An updated literature review. J Cell Physiol 2019;234:5762-74. [Crossref] [PubMed]
- Tait JF, Gibson D. Phospholipid binding of annexin V: effects of calcium and membrane phosphatidylserine content. Arch Biochem Biophys 1992;298:187-91. [Crossref] [PubMed]
- Andree HA, Reutelingsperger CP, Hauptmann R, et al. Binding of vascular anticoagulant alpha (VAC alpha) to planar phospholipid bilayers. J Biol Chem 1990;265:4923-8. [Crossref] [PubMed]
- Wever KE, Wagener FA, Frielink C, et al. Diannexin protects against renal ischemia reperfusion injury and targets phosphatidylserines in ischemic tissue. PLoS One 2011;6:e24276. [Crossref] [PubMed]
- Piët ACA, Post M, Dekkers D, et al. Proximity Ligation Mapping of Microcephaly Associated SMPD4 Shows Association with Components of the Nuclear Pore Membrane. Cells 2022;11:674. [Crossref] [PubMed]
- Schittenhelm L, Hilkens CM, Morrison VL. β2 Integrins As Regulators of Dendritic Cell, Monocyte, and Macrophage Function. Front Immunol 2017;8:1866. [Crossref] [PubMed]
- Tzircotis G, Braga VM, Caron E. RhoG is required for both FcγR- and CR3-mediated phagocytosis. J Cell Sci 2011;124:2897-902. [Crossref] [PubMed]
- Katakai T, Kondo N, Ueda Y, et al. Autotaxin produced by stromal cells promotes LFA-1-independent and Rho-dependent interstitial T cell motility in the lymph node paracortex. J Immunol 2014;193:617-26. [Crossref] [PubMed]
- Varga G, Nippe N, Balkow S, et al. LFA-1 contributes to signal I of T-cell activation and to the production of T(h)1 cytokines. J Invest Dermatol 2010;130:1005-12. [Crossref] [PubMed]
- Jackson JA, Kim EJ, Begley B, et al. Urinary chemokines CXCL9 and CXCL10 are noninvasive markers of renal allograft rejection and BK viral infection. Am J Transplant 2011;11:2228-34. [Crossref] [PubMed]
- Kardos J, Rose TL, Manocha U, et al. Development and validation of a NanoString BASE47 bladder cancer gene classifier. PLoS One 2020;15:e0243935. [Crossref] [PubMed]
- Kurian SM, Velazquez E, Thompson R, et al. Orthogonal Comparison of Molecular Signatures of Kidney Transplants With Subclinical and Clinical Acute Rejection: Equivalent Performance Is Agnostic to Both Technology and Platform. Am J Transplant 2017;17:2103-16. [Crossref] [PubMed]
- Puttarajappa CM, Mehta RB, Roberts MS, et al. Economic analysis of screening for subclinical rejection in kidney transplantation using protocol biopsies and noninvasive biomarkers. Am J Transplant 2021;21:186-97. [Crossref] [PubMed]
- Grewal AS, Friedewald JJ, Abecassis MM. Letter to the AJT Editor re: Puttarajappa et al (doi:
10.1111/ajt.16150 ). Am J Transplant 2021;21:1346-7.10.1111/ajt.16150


