
This article has an erratum available at: http://dx.doi.org/10.21037/tau-2024-02 the article has been update on 2024-10-18 at here.
Single-nucleotide polymorphisms of matrix metalloproteinase genes are associated with graft fibrosis after kidney transplantation
Highlight box
Key findings
• Matrix metalloproteinase (MMP) gene rs9059 promotes the epithelial-mesenchymal transition (EMT) process and is associated with kidney graft fibrosis.
What is known and what is new?
• Fibrosis limits the long-term survival of kidney grafts. Several studies have shown the involvement of MMP genes in allograft tissue injury, including ischemia-reperfusion injury and acute or chronic rejection. However, the role and function of gene polymorphisms in MMPs on interstitial fibrosis formation after kidney transplantation are unknown.
• We provide genetic evidence of the role of MMP9 in the pathogenesis of kidney fibrosis and verified the function of MMP9 mutant promotion to EMT.
What is the implication, and what should change now?
• Our findings provide insights into the etiopathogenesis of renal allograft interstitial fibrosis caused by the MMP gene, which could be used as a candidate target for future treatment.
Introduction
Fibrosis of the transplanted kidney is a major cause of chronic graft loss and limits the long-term survival of kidney grafts (1). The main manifestation of kidney graft fibrosis is interstitial fibrosis and tubular atrophy (IFTA), which is caused by various injuries, including immune rejection, the inflammatory response, and genetic factors (2,3). Recently, the gene polymorphisms that promote the formation of renal fibrosis have been reported in several studies (4-7). Thus, further research needs to be conducted on genetic variables in this field.
Matrix metalloproteinase (MMP) is an endopeptidase in the extracellular matrix (ECM) and contains a variety of proteins, including MMP-1, MMP-2, MMP-3, MMP-7, MMP-9, and MMP-10 (6,8,9). The MMP family plays a critical role in several physiological processes, including organogenesis, tissue wound healing, angiogenesis, cell proliferation, and motility (10). Research has shown that molecules in the MMP family are connected to the development of graft rejection after renal transplantation (11). Rodrigo et al. found that serum MMP-1 may induce acute rejection by regulating the production of the inflammatory cytokines, while MMP-2 and MMP-3 reflect glomerular and interstitial ECM changes in chronic transplant nephropathy (12). In addition, post-transplant MMP-9 concentrations have been reported to be indicators of the long-term function of the transplanted kidney and to be correlated with tubular atrophy and fibrosis (13). The genetic variations within the MMP-3/MMP-12 locus affect the susceptibility of type 1 diabetes patients to diabetic nephropathy (14). A single-nucleotide polymorphism (SNP) analysis of biopsied kidney samples showed that MMP14 is associated with the risk of development of focal segmental glomerulosclerosis (15). However, the role and function of gene polymorphisms in MMPs in interstitial fibrosis formation after kidney transplantation are unknown.
Interstitial fibrosis in transplanted kidneys is mainly caused by the deposition of collagen secreted by fibroblasts. Renal tubular epithelial cells are induced to transform into mesenchymal cells, which become fibroblasts through the process of epithelial-mesenchymal transition (EMT) (16,17). During the EMT process, epithelial cells lose their epithelial phenotype and the expression of junction markers fibronectin (FN) and acquire mesenchymal cell characteristics, such as increased collagen synthesis and expression of α-smooth muscle actin (α-SMA), resulting in ECM protein production (18-20). The effect of SNP in MMP genes on EMT-mediated transplant kidney fibrosis is still not known.
In this study, we used high-throughput and deep next-generation sequencing (NGS) to explore the relationships between MMP gene SNPs in renal transplant recipients and renal allograft fibrosis. The function of MMP9 mutant promotion in EMT was verified by an in-vitro experiment. We present the following article in accordance with the MDAR reporting checklist (available at https://tau.amegroups.com/article/view/10.21037/tau-22-506/rc).
Methods
Ethical statement
The Ethics Committee of Nanjing Medical University’s First Affiliated Hospital reviewed and approved the study protocol (No. 2016-SR-029). By signing the written informed consent form, all the kidney transplant recipients confirmed that they understood the procedures and risks indicated therein. The techniques used in this study adhered to the Declaration of Helsinki (as revised in 2013).
Study population
A total of 200 transplant recipients at the Renal Transplantation Center of The First Affiliated Hospital of Nanjing Medical University who underwent kidney transplantation from February 2010 to December 2015 were recruited for this study. The inclusion and exclusion criteria were described in our previous related study (21). Medical information, including each recipient’s age, sex, weight, demographic details, the cause of the kidney disease, serum creatinine, and human leukocyte antigen (HLA) mismatch, were extracted from hospital records. The immunosuppression regimen after renal transplantation was described in our previous study (22). All the patients were given a 3- or 4-drug immunosuppressive regimen that included cyclosporin A or tacrolimus, mycophenolate mofetil, and prednisone, with or without sirolimus. The drug doses were modified based on the therapeutic drug monitoring results of serum creatinine levels. Basiliximab or anti-human thymocyte immunoglobulin was used in the transplant induction therapy.
The transplant recipients participating in this study provided their informed consent to undergo a post-renal puncture biopsy, and the severity and type/grade of the fibrosis were diagnostically classified and scored by a nephrologist according to the Banff 2017 criteria for allografts.
Sample preparation and target sequencing analysis
Peripheral blood (2 mL) samples were obtained from each participant, and DNA was extracted. The content and purity of the genomic DNA (gDNA) were quantified, and gene integrity was confirmed by agarose gel electrophoresis. A pool containing upstream and downstream oligonucleotides unique to the selected areas of interest was hybridized into the gDNA samples. With an ABI 9700 polymerase chain reaction (PCR) apparatus, quantitative detection was conducted, and fragmentation was followed by end repair, dA tailing, and sequencing adaptor ligation. The adapter-ligated DNA was amplified using selective, limited-cycle PCR and then quantified. The mixture was amplified for 16 PCR cycles and quantified. The collected libraries were denatured and placed at 12 to 16 pmol/L onto an Illumina cBot instrument for cluster creation according to the manufacturer’s recommendations. Each high-quality sequence (HiSeq) lane could sequence up to 20 WUCaMP libraries. To analyze the sequencing data for the human reference sequence UCSC hg19 (NCBI build 37.2) assembly, we used the Genome Analysis Tool Kit, Picard Software, and dbSNP 132. The MuTect 1.1.5 and VarScan 2.3.6. programs were used to identify the potential somatic variant cells.
In-vitro experiment
The human kidney 2 (HK2) cells were cultured in Dulbecco’s Modified Eagle Medium/F12 media with 10% fetal bovine serum and 1% penicillin-streptomycin in a humidified environment containing 5% carbon dioxide. The cells were serum-starved overnight and then treated with 10 ng/mL of transforming growth factor-β (TGF-β) for 0, 12, 24, or 48 hours to determine the effect of TGF-β on the HK2 cells. Total protein and RNA were extracted for the western blot assays and quantitative real-time PCR (qRT-PCR). The protocols for the western blot assays and qRT-PCR were detailed in previous research (19). The following primary antibodies were used: anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (1:1,000; CST, USA), anti-a-SMA (1:1,000; Abcam, USA), anti-CD31 (1:1,000; CST), anti-FN (1:1,000; BD Biosciences, USA), and anti-MMP9 (1:1,000; Abcam).
Wound-healing tests and transwell assays
The wound-healing tests were carried out by plating the cells in 6-well culture plates. Pipette tips were used to scrape the cells, which were then rinsed in phosphate-buffered saline. Fresh whole-media containing TGF-β (10 ng/mL) was introduced to the scratched cells for 24 hours. Images were taken after scraping the cells at ×100 magnification for 0 and 24 h under an inverted microscope. Each assay was repeated independently at least 3 times.
The cells were cultured on 24-well culture plates to study HK2 migration. For the number of cells per top chamber, the cells were injected at a density of 5×104 on the upper surface of polycarbonate filters with 8-µm holes in media containing 10 ng/mL of TGF-β. The non-invasive cells on the top chamber surface were removed using a cotton swab after 48 hours of incubation at 37 ℃. The cells on the bottom were fixed with 4% paraformaldehyde and then stained with 0.1% crystal violet for 10 min. Using a phase contrast microscope at ×100 magnification, the number of cells present on the bottom surface was counted to quantify cell migration.
Statistical analysis
The Hardy-Weinberg equilibrium (HWE) analysis was performed using the RStudio version 4.0.5 software (Boston, MA, USA) and the “SNPassoc” package version 2.0-2, while the minor allele frequencies (MAFs) and linkage disequilibrium (LD) blocks were performed using the Haploview version 4.2 program (Broad Institute, Cambridge, MA, USA). General linear models (GLMs) were used to investigate the possible link between the clinical factors and renal allograft fibrosis. Five inheritance models were used in the genotype association analysis, including the dominant model (minor allele homozygotes plus heterozygotes vs. major allele homozygotes), the recessive model (minor allele homozygotes vs. heterozygotes plus major homozygotes), the codominant model (major allele homozygotes vs. heterozygotes vs. minor allele homozygotes), and the overdominant model (version 2.0-2).
GraphPad Prism 7.0 software was used to calculate the mean and standard deviation (SD) of at least 3 separate trials. An unpaired Student’s t-test or 2-way analysis of variance was used to compare the mutant and wild-type groups. A P value <0.05 was considered statistically significant. When multiple comparisons occurred, Bonferroni correction procedures were used to avoid inflating the P values.
Results
Clinical characteristics of the involved patients
Based on the occurrence of biopsy-proven transplanted kidney fibrosis events, the patients were classified into the graft fibrosis group (n=69) and the control group (n=131). In this study cohort, there were no confounding factors, such as age, recipient gender, type of donor, and HLA matching (P>0.05). All the patients were undergoing primary kidney transplantation with negative panel reactive antibody pre-surgery. Approximately 12% of the patients were treated with sirolimus postoperatively. Detailed demographic information has been provided in the supplementary document (Table S1).
The degree of IFTA was identified and classified as mild, moderate, or severe, and the Banff score in each biopsy was obtained by 2 independent pathologists (23). As Table 1 shows, the number of patients classified as mild, moderate, or severe was 32 (46.38%), 24 (34.78%), and 13 (18.84%), respectively.
Table 1
| Variables | Values |
|---|---|
| Fibrosis group case number | 69 |
| IFTA | |
| Mild | 32 |
| Moderate | 24 |
| Severe | 13 |
| Banff score* (mean ± SD) | |
| CG | 0.072±0.31 |
| CI | 1.43±0.53 |
| CT | 1.57±0.67 |
| CV | 0.57±0.79 |
| CH | 0.13±0.38 |
*, Banff score was obtained following the Banff 2017 revised diagnostic criteria. IFTA, interstitial fibrosis and tubular atrophy; SD, standard deviation; CG, glomerular basement membrane double contours; CI, interstitial fibrosis; CT, tubular atrophy; CV, vascular fibrous intimal thickening; CH, interstitial hemorrhage.
Identification of the SNPs in the MMP genes
We performed target sequencing based on NGS for the MMP genes and identified 41 SNPs involved in 5 related genes (i.e., MMP1, MMP2, MMP3, MMP9, and MMP10). Table S2 lists the genetic information of the 5 identified SNPs.
Next, the common variants were defined as those with an MAF >0.05, and a threshold of 0.05 was set for the HWE values. After performing the MAF and HWR analysis, 41 SNPs were identified. Further, 20 tagger SNPs were identified by the LD analysis (Figure 1), including rs9509, rs17859975, Chr11:102711338, rs243835, rs11640428, rs13969, Chr11:102713046, rs12599775, rs243846, rs243849, rs1030868, rs2250889, rs17860949, rs1053605, rs14070, rs3918251, rs470558, rs1132896, rs171498, and rs470263.
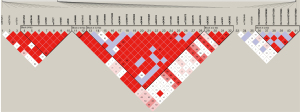
Multiple inheritance model analysis
In relation to the continuous variables, we included the indicators in a sequential manner in a model that used the adjusted confounding factors based on the relative degree of their connection with allograft fibrosis. In our previous study, the administration of sirolimus was identified as the confounding factor by the GLM analysis, which suggests that sirolimus treatment significantly affects the severity of fibrosis in renal grafts (21).
Next, after using the Bonferroni correction to adapt the various sirolimus treatments (adjusted P=0.005), a multiple inheritance model analysis was performed for each of the 5 analytical models (i.e., the dominant, recessive, additive, overdominant, and codominant models) to investigate the association between these MMP tagger SNPs and renal graft fibrosis after kidney transplantation (Table S3). The results showed that 4 SNPs (i.e., rs243846, rs1053605, rs9509, and rs11640428) were significantly correlated with allograft fibrosis in at least 1 or more of the models (Table 2).
Table 2
| SNPs | Codominant | Dominant | Recessive | Overdominant | Log-additive |
|---|---|---|---|---|---|
| rs243846 | 0.041 | 0.14 | 0.015 | 0.51 | 0.019 |
| rs1053605 | 0.073 | 0.022 | 0.62 | 0.028 | 0.028 |
| rs9509 | 0.034 | 0.13 | 0.015 | 0.52 | 0.038 |
| rs11640428 | 0.0090 | 0.0064 | 0.71 | 0.0023 | 0.050 |
SNPs, single-nucleotide polymorphisms.
We also explored the effect of the SNPs on IFTA and found that rs9509 was significantly associated with the severity of IFTA (likelihood ratio =20.08, P<0.0001). No positive results were observed in the other significant SNPs (Table S4). We also chose 3 genotypes (i.e., CC, TC, and TT) and compared the variations among various degrees of IFTA. We found no significant differences in the degrees of IFTA (i.e., mild, moderate, and severe IFTA) among the CC, CT, and TT genotypes. Notably, we discovered that when 1 T allele occurred (in the TC genotype), there was a slight increase in the severity of IFTA as compared to the group in which the T allele did not occur. Additionally, there was a marked increase in the proportion of severe IFTA in samples with 2 T alleles (in the TT genotypes). These findings suggested that as the frequency of T alleles increased, so too did the severity of IFTA.
The MMP9 rs9509 mutation promotes EMT in HK2 cells
TGF-β is recognized as an effective inducer of kidney fibrosis and has been used to stimulate the EMT in previous research (19). To investigate the relationship between MMP9 and EMT in renal interstitial fibrosis, we conducted an in-vitro experiment dependent on TGF-β induction. Figure S1 shows the results of the construction and verification of the MMP9 rs9509 mutant plasmid. The HK2 cells were stimulated with 10 ng/mL of TGF-β for different times (0, 12, 24, or 48 hours). TGF-β increased the protein expression of MMP9 in a time-dependent manner (Figure 2A). These data showed that the HK-2 cells underwent transition into mesenchymal cells after the administration of TGF-β, and MMP9 was involved.
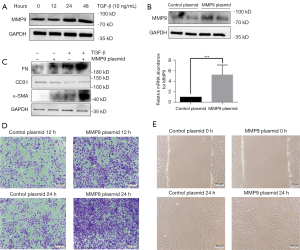
Next, to determine the biological significance of the kidney fibrosis-associated rs9059 mutation in the MMP9 gene, we transfected the rs9059 mutated plasmids into HK-2 cells (with MMP9 wild-type cells as the control). We found that the expression of MMP9 protein and messenger RNA was significantly increased by the transfer of the rs9059 mutation in the HK-2 cells (Figure 2B). The western blot assay results demonstrated that the expression of FN, CD31, and α-SMA was significantly increased, while the expression of CD31 was significantly reduced in the rs9059 plasmids transferred cells after the TGF-β stimulation (Figure 2C). In addition, wound-healing tests and transwell assays were carried out to investigate the development of migratory potential following mutant therapy. The results demonstrated that the cells transfected with the rs9059 mutant plasmid exhibited more mobility and migratory capacity than the control wild-type cells (Figure 2D,2E). Taken together, these data suggest that the rs9059 mutation of the MMP9 gene further facilitated EMT.
Discussion
In solid organ transplantation, several studies have shown the involvement of MMP in allograft injury, including ischemia-reperfusion injury, and acute or chronic rejection (24,25). Further research should be conducted on the role of MMP genes in transplanted kidney fibrosis. In this study, we explored the association between SNPs in the MMP genes and fibrosis in kidney transplant recipients. MMP9 rs9059 was found to be significantly correlated with allograft renal fibrosis.
Many studies of renal graft fibrosis in recent years have provided evidence of the important role of SNPs in this process. Caveolin-1 rs4730751 SNP has been reported to be significantly associated with allograft failure in independent cohorts (26). Another study found that the presence of the donor SHROOM3 locus rs17319721 allele is associated with increased SHROOM3 expression and fibrosis in homozygous grafts, which was confirmed in vitro (4). The SNPs of TGFβ1 involving promoter regions show more decisive associations with the progression of transplanted kidney fibrosis compared to healthy control recipients (27).
Chronic kidney transplant injury is characterized by IFTA and the accumulation of ECM. MMPs and their inhibitors, which are regulators of ECM physiology, cause ECM deposition and promote organ fibrosis when the dynamic balance is disrupted. Conversely, the activation of MMPs can form a cascade effect to promote the amplification of MMP family protein synergy. Research has shown that MMPs can induce EMT in vivo, they can act on laminin and type IV collagen in the tubular basement membrane, disrupting the integrity of the basement membrane and allowing transformed mesenchymal cells to migrate and invade the renal mesenchyme (28,29). The results of our in-vitro studies showed that MMP9 mutations significantly contribute to the progression of EMT in HK-2 cells, as evidenced by altered expression levels of marker proteins and increased cell invasive migration ability, which is consistent with the previous results (28,29).
In our study, we sequenced locus mutations in the MMP genes and identified an SNP mutation rs9059 that may contribute to graft fibrosis. Natural sequence variations, such as non-synonymous SNPs in the promoters and coding regions of MMP genes, may cause different expressions of MMPs in different people (30). Similarly, we found that the rs9059 mutation positively affects MMP9 protein expression in vitro. These findings explain the results observed in our cohort. We found that as the T alleles increase, the pathological manifestations of IFTA are more severe, which suggests that the difference is meaningful and should be investigated further with a larger sample size. Thus, we identified a specific biomarker for the diagnosis of graft fibrosis at the early stage post-kidney transplantation to predict the long-term prognosis of the transplanted kidney. In addition, we found that this locus may represent a prospective therapeutic target whereby MMP9 and its nucleotide mutations can be targeted to optimize the long-term prognosis of the transplanted kidney.
This study had several limitations. First, only 200 participants were enrolled in this study, which may have created a potential bias in the final outcomes. Second, there was a lack of a second cohort to validate the results derived from the primary exploration cohort, but the in-vitro experiment results provide further evidence to support the conclusions drawn. Thus, a prospective, well-designed study should be carried out for further validation, and the interaction between these SNPs in kidney graft fibrosis need to be examined further in future studies.
Conclusions
MMP9 is involved in the pathogenesis of kidney fibrosis after renal transplantation, and the SNP rs9059 mutant is significantly connected to remarkable kidney allograft pathological changes and can promote the process of EMT. Our findings provide insights into the etiopathogenesis of renal allograft interstitial fibrosis caused by SNP variations of MMP genes and MMP9 could be used as a candidate target for future treatment.
Acknowledgments
Funding: This work was supported by the National Natural Science Foundation of China (Nos. 82170769, 82070769, 81900684, 81870512, and 81770751), the Project of Jiangsu Province for Important Medical Talent (No. ZDRCA2016025), the “333 High-Level Talents Project” in Jiangsu Province (Nos. BRA2017532 and BRA2016514), the Standardized Diagnosis and Treatment Research Program of Key Diseases in Jiangsu Province (No. BE2016791), the Open Project Program of the Health Department of Jiangsu Province (No. JSY-2-2016-099), and the Jiangsu Province Natural Science Foundation Program (No. BK20191063).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://tau.amegroups.com/article/view/10.21037/tau-22-506/rc
Data Sharing Statement: Available at https://tau.amegroups.com/article/view/10.21037/tau-22-506/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tau.amegroups.com/article/view/10.21037/tau-22-506/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of Nanjing Medical University’s First Affiliated Hospital (No. 2016-SR-029) and informed consent was obtained from all the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Boor P, Floege J. Renal allograft fibrosis: biology and therapeutic targets. Am J Transplant 2015;15:863-86. [Crossref] [PubMed]
- Sellarés J, de Freitas DG, Mengel M, et al. Understanding the causes of kidney transplant failure: the dominant role of antibody-mediated rejection and nonadherence. Am J Transplant 2012;12:388-99. [Crossref] [PubMed]
- Nankivell BJ, Shingde M, Keung KL, et al. The causes, significance and consequences of inflammatory fibrosis in kidney transplantation: The Banff i-IFTA lesion. Am J Transplant 2018;18:364-76. [Crossref] [PubMed]
- Menon MC, Chuang PY, Li Z, et al. Intronic locus determines SHROOM3 expression and potentiates renal allograft fibrosis. J Clin Invest 2015;125:208-21. [Crossref] [PubMed]
- Yang B, Feng W, Li Y, et al. Interleukin 18 -607 A/C Gene Polymorphism is Associated With Susceptibility to IgA Nephropathy in a Chinese Han Population. Appl Immunohistochem Mol Morphol 2017;25:725-30. [Crossref] [PubMed]
- Bassiouni W, Ali MAM, Schulz R. Multifunctional intracellular matrix metalloproteinases: implications in disease. FEBS J 2021;288:7162-82. [Crossref] [PubMed]
- Jiang X, Konkalmatt P, Yang Y, et al. Single-nucleotide polymorphisms of the dopamine D2 receptor increase inflammation and fibrosis in human renal proximal tubule cells. Hypertension 2014;63:e74-80. [Crossref] [PubMed]
- Mahalanobish S, Saha S, Dutta S, et al. Matrix metalloproteinase: An upcoming therapeutic approach for idiopathic pulmonary fibrosis. Pharmacol Res 2020;152:104591. [Crossref] [PubMed]
- Fischer T, Riedl R. Challenges with matrix metalloproteinase inhibition and future drug discovery avenues. Expert Opin Drug Discov 2021;16:75-88. [Crossref] [PubMed]
- Mittal R, Patel AP, Debs LH, et al. Intricate Functions of Matrix Metalloproteinases in Physiological and Pathological Conditions. J Cell Physiol 2016;231:2599-621. [Crossref] [PubMed]
- Inkinen KA, Soots AP, Krogerus LA, et al. Fibrosis and matrix metalloproteinases in rat renal allografts. Transpl Int 2005;18:506-12. [Crossref] [PubMed]
- Rodrigo E, López-Hoyos M, Escallada R, et al. Circulating levels of matrix metalloproteinases MMP-3 and MMP-2 in renal transplant recipients with chronic transplant nephropathy. Nephrol Dial Transplant 2000;15:2041-5. [Crossref] [PubMed]
- Kwiatkowska E, Domanski L, Bober J, et al. Urinary Metalloproteinases-9 and -2 and Their Inhibitors TIMP-1 and TIMP-2 are Markers of Early and Long-Term Graft Function After Renal Transplantation. Kidney Blood Press Res 2016;41:288-97. [Crossref] [PubMed]
- Kure M, Pezzolesi MG, Poznik GD, et al. Genetic variation in the matrix metalloproteinase genes and diabetic nephropathy in type 1 diabetes. Mol Genet Metab 2011;103:60-5. [Crossref] [PubMed]
- Munkert A, Helmchen U, Kemper MJ, et al. Characterization of the transcriptional regulation of the human MT1-MMP gene and association of risk reduction for focal-segmental glomerulosclerosis with two functional promoter SNPs. Nephrol Dial Transplant 2009;24:735-42. [Crossref] [PubMed]
- Lovisa S, LeBleu VS, Tampe B, et al. Epithelial-to-mesenchymal transition induces cell cycle arrest and parenchymal damage in renal fibrosis. Nat Med 2015;21:998-1009. [Crossref] [PubMed]
- Gui Z, Suo C, Tao J, et al. Everolimus Alleviates Renal Allograft Interstitial Fibrosis by Inhibiting Epithelial-to-Mesenchymal Transition Not Only via Inducing Autophagy but Also via Stabilizing IκB-α. Front Immunol 2022;12:753412. [Crossref] [PubMed]
- Krieg T, Abraham D, Lafyatis R. Fibrosis in connective tissue disease: the role of the myofibroblast and fibroblast-epithelial cell interactions. Arthritis Res Ther 2007;9:S4. [Crossref] [PubMed]
- Wang Z, Han Z, Tao J, et al. Role of endothelial-to-mesenchymal transition induced by TGF-β1 in transplant kidney interstitial fibrosis. J Cell Mol Med 2017;21:2359-69. [Crossref] [PubMed]
- Zhou J, Cheng H, Wang Z, et al. Bortezomib attenuates renal interstitial fibrosis in kidney transplantation via regulating the EMT induced by TNF-α-Smurf1-Akt-mTOR-P70S6K pathway. J Cell Mol Med 2019;23:5390-402. [Crossref] [PubMed]
- Liu X, Liu K, Gui Z, et al. Single Nucleotide Polymorphisms of IL-33 Gene Correlated with Renal Allograft Fibrosis in Kidney Transplant Recipients. J Immunol Res 2021;2021:8029180. [Crossref] [PubMed]
- Wang Z, Zheng M, Yang H, et al. Association of Genetic Variants in CYP3A4, CYP3A5, CYP2C8, and CYP2C19 with Tacrolimus Pharmacokinetics in Renal Transplant Recipients. Curr Drug Metab 2019;20:609-18. [Crossref] [PubMed]
- Haas M, Loupy A, Lefaucheur C, et al. The Banff 2017 Kidney Meeting Report: Revised diagnostic criteria for chronic active T cell-mediated rejection, antibody-mediated rejection, and prospects for integrative endpoints for next-generation clinical trials. Am J Transplant 2018;18:293-307. [Crossref] [PubMed]
- Campbell LG, Ramachandran S, Liu W, et al. Different roles for matrix metalloproteinase-2 and matrix metalloproteinase-9 in the pathogenesis of cardiac allograft rejection. Am J Transplant 2005;5:517-28. [Crossref] [PubMed]
- Falk V, Soccal PM, Grünenfelder J, et al. Regulation of matrix metalloproteinases and effect of MMP-inhibition in heart transplant related reperfusion injury. Eur J Cardiothorac Surg 2002;22:53-8. [Crossref] [PubMed]
- Moore J, McKnight AJ, Simmonds MJ, et al. Association of caveolin-1 gene polymorphism with kidney transplant fibrosis and allograft failure. JAMA 2010;303:1282-7. [Crossref] [PubMed]
- Tampe B, Zeisberg M. Contribution of genetics and epigenetics to progression of kidney fibrosis. Nephrol Dial Transplant 2014;29:iv72-9. [Crossref] [PubMed]
- Tan TK, Zheng G, Hsu TT, et al. Macrophage matrix metalloproteinase-9 mediates epithelial-mesenchymal transition in vitro in murine renal tubular cells. Am J Pathol 2010;176:1256-70. [Crossref] [PubMed]
- Liu Y. Epithelial to mesenchymal transition in renal fibrogenesis: pathologic significance, molecular mechanism, and therapeutic intervention. J Am Soc Nephrol 2004;15:1-12. [Crossref] [PubMed]
- Hu Z, Huo X, Lu D, et al. Functional polymorphisms of matrix metalloproteinase-9 are associated with risk of occurrence and metastasis of lung cancer. Clin Cancer Res 2005;11:5433-9. [Crossref] [PubMed]


